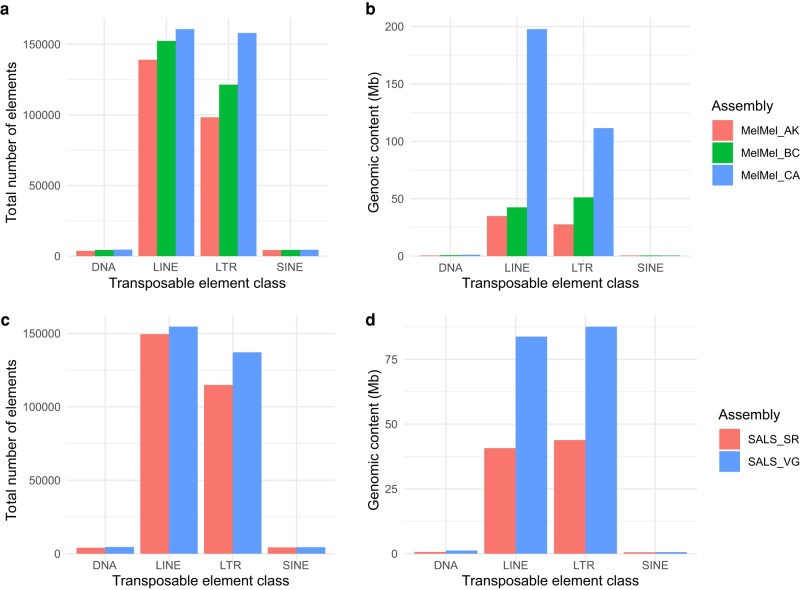
Fig. 4.
a to b) Comparison of transposable element annotation across three song sparrow (Melospiza melodia) assemblies. a) Total number of transposable elements found in each assembly. b) Total genomic content (Mb) of transposable element content identified in each of the three song sparrow assemblies. MelMel_AK (red) is an assembly from an Alaskan bird sequenced using short-read and Chicago library technology. MelMel_BC (green) is a bird from British Columbia sequenced using Illumina short-read and PacBio SMRT long-read approaches MelMel_CA was generated using Hi-C and PacBio long-read approaches for this paper. c and d) Comparison of TE annotations in two saltmarsh sparrow assemblies (Ammospiza caudacuta). (c) Total number of transposable elements found in each assembly. b) Total genomic content (Mb) of transposable element content identified in each of the three song sparrow assemblies. SALS_SR (Red) was assembled from Illumina short reads and the SALS_VG (blue) assembly was assembled using PacBio long-read and Omni-C approaches with the Vertebrate Genomics Project pipeline.