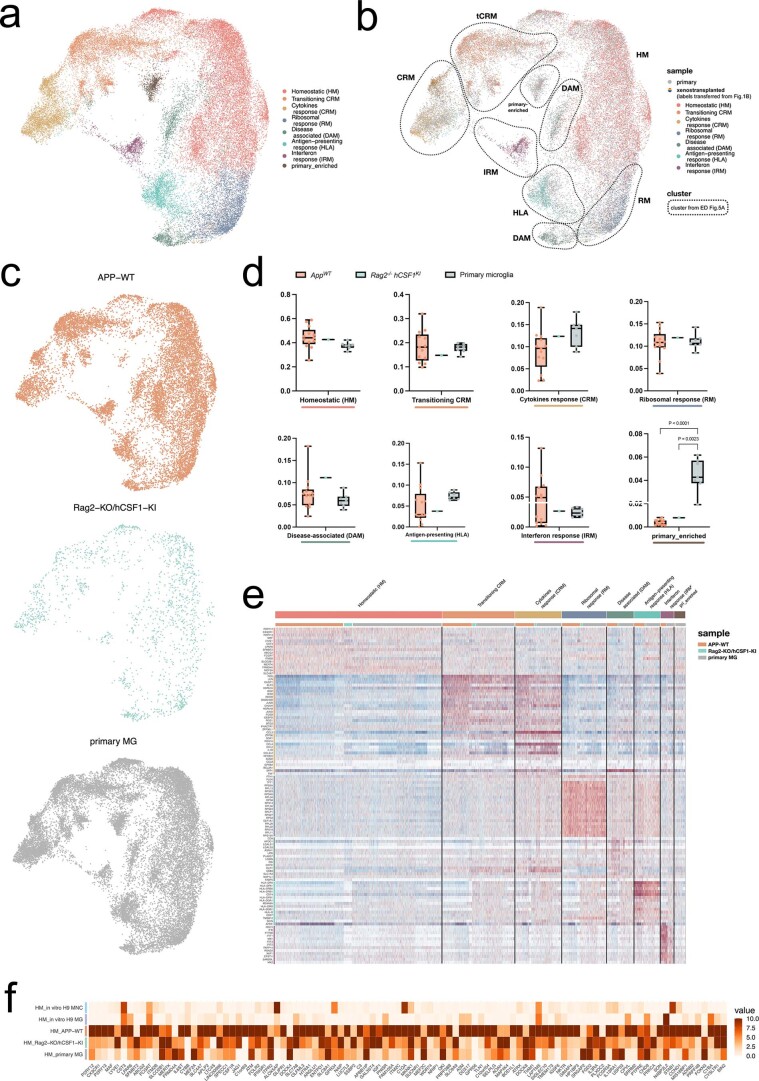
Extended Data Fig. 1. Comparison of human microglia transplanted in AppWT and Rag2−/−hCSFKI mice, and primary microglia isolated from human surgical resections published before14.
a, UMAP plot of the 39,538 cells passing quality control, coloured by annotated cell states after removal of CAMs, other myeloid, low quality and proliferating clusters. b, UMAP plots as in a, coloured by sample. c, UMAP plots as in a, split between the three experimental groups. d, Box plot showing proportion of cells across all identified clusters (box plots are limited by lower and upper quartiles and midline indicates median; whiskers show minimum to maximum value. Each dot represents a single replicate, n = 13 for AppWT, n = 1 for Rag2−/−/hCSF1KI, n = 7 for primary MG. One-way ANOVA with Tukey’s multiple comparisons as post-hoc test, alpha = 0.05, significance was set as P-adjusted value < 0.05). e, Heatmap displaying the top20 most upregulated genes in each cluster as in a. f, Heatmap depicting the expression levels of homeostatic microglial markers across all experimental groups (normalized expression scaled by gene is shown).