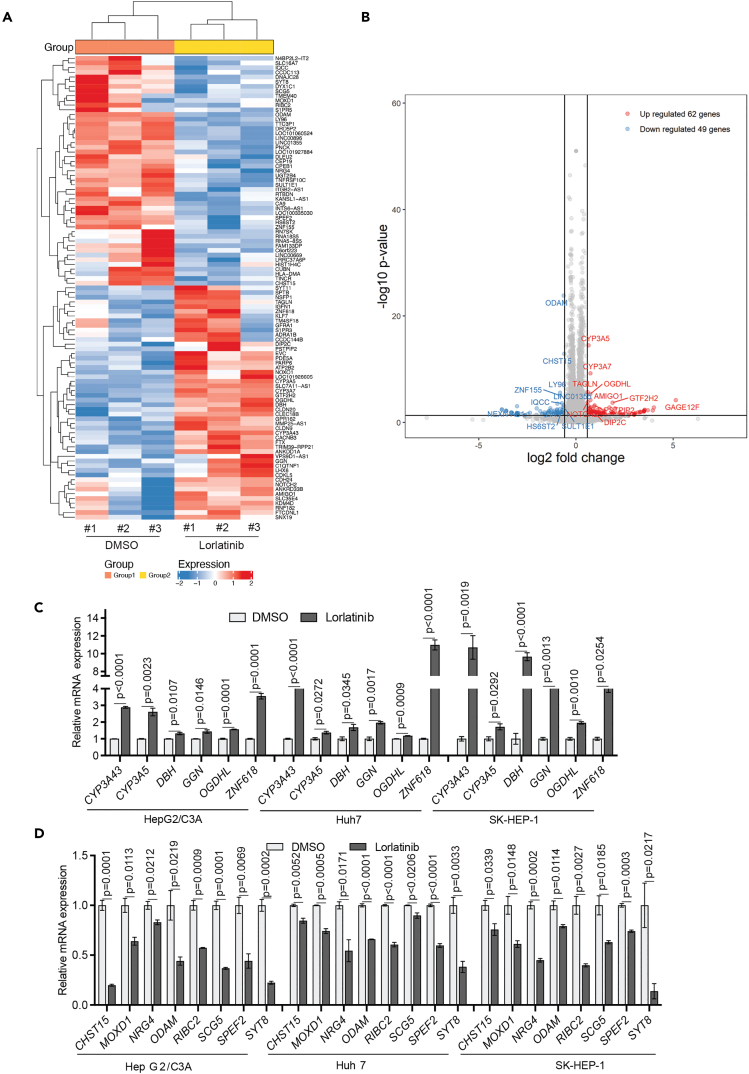
Figure 6.
Transcriptome-wide mRNA expression profiling identifies Lorlatinib-responsive gene signature in HCC cells
(A and B) HepG2/C3A cells were treated with either DMSO, or 1 μM lorlatinib for 24 h, after which RNA sequencing was performed. Heatmap (A) showing the top 100 most differentially regulated genes in lorlatinib treatment compared to DMSO treatment is shown. Volcano plot (B) showing differentially expressed genes in lorlatinib compared to DMSO-treated HepG2/C3A cells.
(C) mRNA expression for indicated genes that were identified to be upregulated from RNA-seq measured by RT-qPCR in HepG2/C3A, Huh7, and SK-HEP-1 cells after treatment with lorlatinib compared to cells treated with DMSO. ACTINB mRNA expression was used as an internal normalization control. Relative mRNA expression (n = 3 each) for indicated genes is plotted. For HepG2/C3A for CYP3A43 gene p < 0.0001, t = 36.71, df = 4, CYP3A5 gene p = 0.0023, t = 6.907, df = 4, DBH gene p = 0.0107, t = 4.516, df = 4, GGN gene p = 0.0146, t = 4.120, df = 4, OGDHL gene p = 0.0001, t = 14.57, df = 4, ZNF618 gene p = 0.0001, t = 15.25, df = 4. For Huh7 for CYP3A43 gene p < 0.0001, t = 18.40, df = 4, CYP3A5 gene p = 0.0272, t = 3.403, df = 4, DBH gene p = 0.0345, t = 3.150, df = 4, GGN gene p = 0.0017, t = 7.508, df = 4, OGDHL gene p = 0.0009, t = 8.744, df = 4, ZNF618 gene p < 0.0001, t = 17.81, df = 4. For SK-HEP-1 for CYP3A43 gene p = 0.0019, t = 7.315, df = 4, CYP3A5 gene p = 0.0292, t = 3.328, df = 4, DBH gene p < 0.0001, t = 15.66, df = 4, GGN gene p = 0.0013, t = 8.063, df = 4, OGDHL gene p = 0.0010, t = 8.518, df = 4, ZNF618 gene p = 0.0254, t = 3.477, df = 4. p values were calculated using a two-tailed, unpaired Student’s t test.
(D) mRNA expression for indicated genes that were identified to be downregulated from RNA-seq measured by RT-qPCR in HepG2/C3A, Huh7, and SK-HEP-1 cells after treatment with lorlatinib compared to cells treated with DMSO. ACTINB mRNA expression was used as an internal normalization control. Relative mRNA expression (n = 3 each) for indicated genes is plotted. For HepG2/C3A for CHST15 gene p = 0.0001, t = 15.54, df = 4, MOXD1 gene p = 0.0113, t = 4.448, df = 4, NRG4 gene p = 0.0212, t = 3.683, df = 4, ODAM gene p = 0.0219, t = 3.646, df = 4, RIBC2 gene p = 0.0009, t = 8.759, df = 4, SCG5 gene p = 0.0001, t = 14.89, df = 4, SPEF2 gene p = 0.0069, t = 5.120, df = 4, SYT8 gene p = 0.0002, t = 12.73, df = 4. For Huh7 for CHST15 gene p = 0.0052, t = 5.548, df = 4, MOXD1 gene p = 0.0005, t = 10.59, df = 4, NRG4 gene p = 0.0171, t = 3.930, df = 4, ODAM gene p < 0.0001, t = 22.11, df = 4, RIBC2 gene p < 0001, t = 16.45, df = 4, SCG5 gene p = 0.0206, t = 3.711, df = 4, SPEF2 gene p < 0.0001, t = 17.52, df = 4, SYT8 gene p = 0.0033, t = 6.274, df = 4. For SK-HEP-1 for CHST15 gene p = 0.0339, t = 3.167, df = 4, MOXD1 gene p = 0.0148, t = 4.103, df = 4, NRG4 gene p = 0.0002, t = 13.78, df = 4, ODAM gene p = 0.0114, t = 4.438, df = 4, RIBC2 gene p = 0.0027, t = 6.639, df = 4, SCG5 gene p = 0.0185, t = 3.838, df = 4, SPEF2 gene p = 0.0003, t = 11.80, df = 4, SYT8 gene p = 0.0217, t = 3.654, df = 4. p values were calculated using a two-tailed, unpaired Student’s t test. All quantitative data represent the mean ± SEM; see also, Figures S3 and S4 and Table S1.