Figure 5. Modeling, concentration estimates and in vitro microrheology suggest rRNA-driven changes to nucleolar rheology.
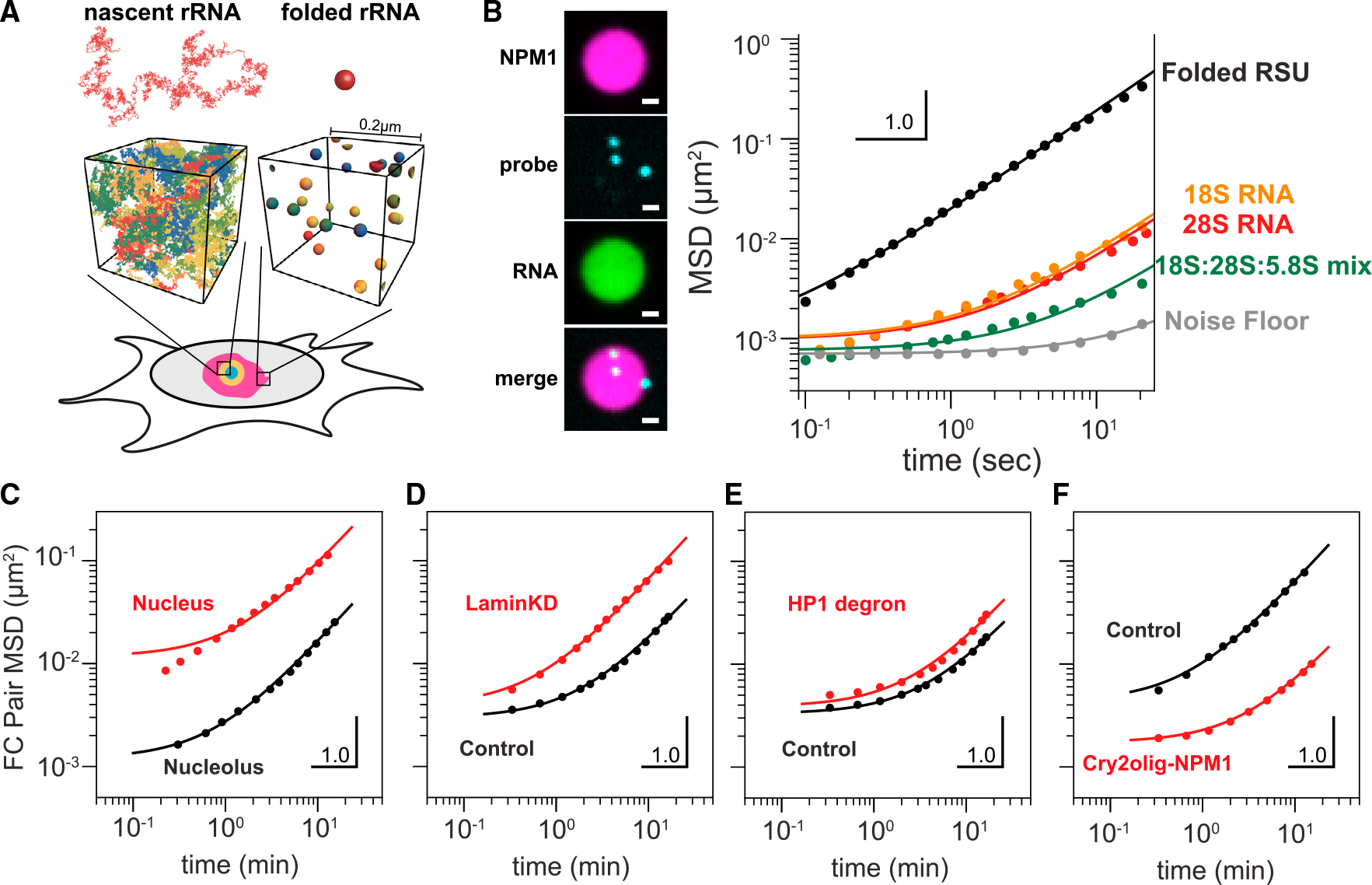
(A) Modeled as a random coil, nascent rRNA is ~15-fold larger in diameter than a folded rRNA subunit, suggesting substantial degree of entanglement between nascent chains, but not folded RSUs, using rRNA transcript density estimated from experimental measurements (supplemental information).
(B) NPM1 (magenta) and ribosomal RNAs (green) were combined to generate in vitro NPM1+rRNA droplets. MSD of microrheological probes in NPM1+rRNA in vitro droplets. E. coli rRNAs (black) are purified from mature polysomes. Orange: in vitro transcribed 18S rRNA. Red: in vitro transcribed 28S rRNA. Green: equimolar mixture of all three in vitro transcribed 18S, 28S, and 5.8S RNAs. Gray: noise floor.
(C) Pairwise MSD of long-time dynamics of FCs, labeled by RPA16-GFP, in HEK cells (“nucleolus”; 508 pairs of FCs in seven nucleoli) and engineered droplets in U2OS nuclei (“nucleoplasm”; 31,582 pairs in 18 cells, re-analyzed data from.37 FCs fit a nonlinear Maxwell model, whereas engineered droplets do not.
(D and E) (D) MSD comparing FC dynamics in control vs. Lamin knockdown cells and (E) HP1 degron cells without (black) and with (red) auxin treatment.
(F) MSD comparing control cells with Cry2olig-NPM1-activated cells.
