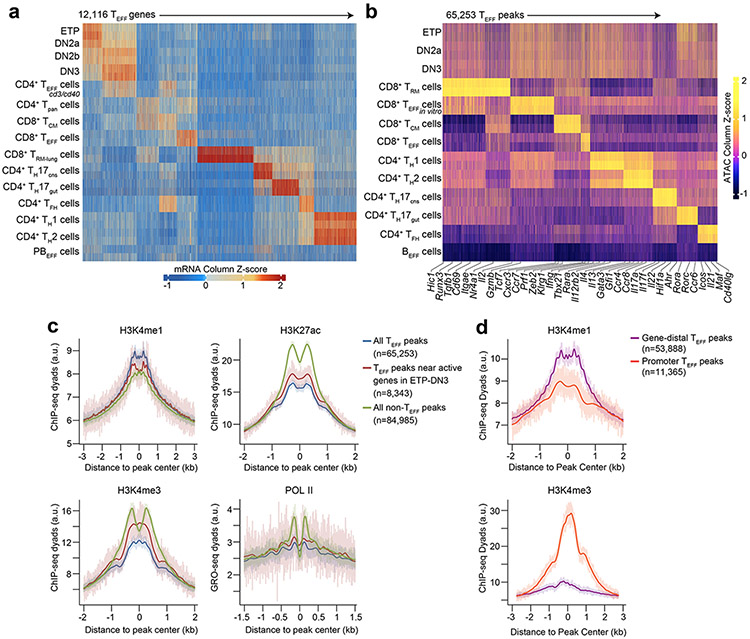
Extended Data Figure 4. Enhancer priming mediates early chromatin accessibility at TEFF loci.
a, Heatmap of Z scores of TPM-normalized RNA-seq reads across the k-means clustered (k=10) transcriptomes (less genes robustly expressed in PBEFF cells) of ETP, DN2a, Lin−c-KitintCD25hiCD44hiCD4−CD8− (DN2b), DN3, CD4+ TEFF cd3/cd40, CD4+ Tpan, TCM, CD8+ TEFF, TRM, TH17CNS, TH17gut, TFH, TH1 and TH2 cells (n=2 for each). b, Heatmap of Z scores of TSS-normalized ATAC-seq fragments across the k-means clustered (k=9) TEFF loci in ETP, DN2, DN3, TRM, CD8+ TEFF in vitro, CD8+ TEFF, TCM, TH1, TH2, TH17CNS, TH17gut, TFH cells and BEFF cells (n=2 each). Labeled columns highlight ATAC-seq peaks whose nearest gene represents a canonical gene reflective of a distinct T effector program. c, Representative aggregate histograms of H3K4me131 (1 of n=2), H3K4me331 (1 of n=2), H3K27ac32 (1 of n=2) or RNA Polymerase II31 (POL II) (1 of n=2) ChIP-seq or GRO-seq dyads intersecting all TEFF loci (blue), TEFF loci near active genes in ETP-DN3 cells (blue) and all non-TEFF loci (green) in DN2-DN3 thymocytes. d, Representative aggregate histograms of H3K4me131 (1 of n = 2), H3K4me331 (1 of n = 2) ChIP-seq dyads at promoter vs. gene-distal TEFF loci in DN2-DN3 thymocytes.