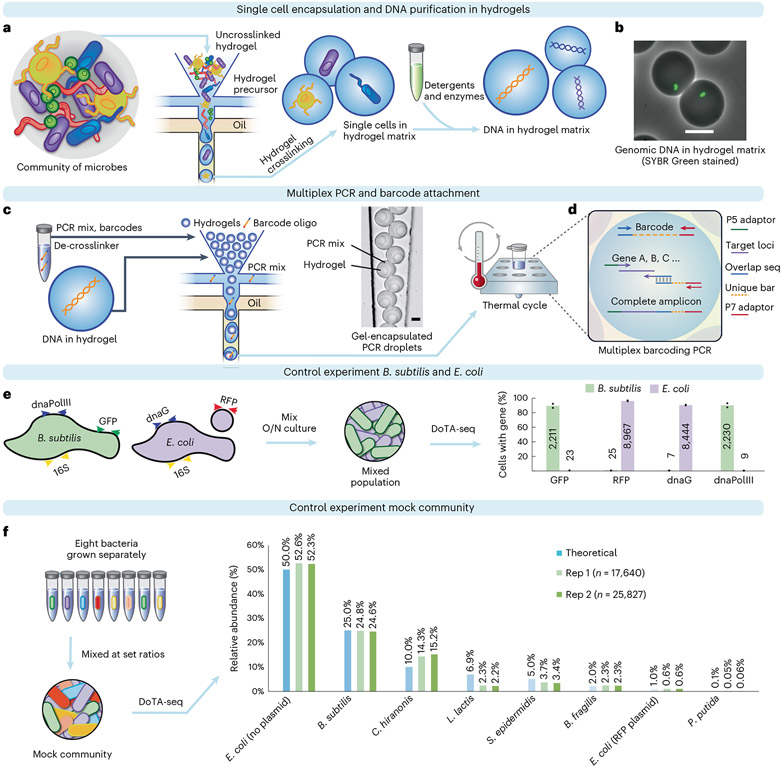
Fig. 1 ∣. DoTA-seq profiles single-cell genetic loci in Gram-negative and -positive bacteria.
a, Overview of the DoTA-seq workflow. b, Fluorescent and brightfield image overlay showing lysed of B. subtilis cells inside hydrogels. SYBR Green staining reveals the trapped genomic DNA inside the hydrogels, whereas the lysed cell is no longer visible in brightfield. Scale bar, 20 μm. For more images of the complete lysis workflow, see Extended Data Fig. 1. c, Hydrogels are re-encapsulated with a DoTA-seq PCR mix with unique barcode oligonucleotides at a limiting dilution. Microscopy image shows a representative image of the re-encapsulated gels at the outlet of the microfluidic device. Scale bar, 20 μm. Oligo, oligonucleotide. d, Schematic of the barcoding multiplex PCR reaction inside droplets. Seq, sequencing. e, Grouped bar plot by gene shows the fraction of cells with the given gene detected in B. subtilis (green bars, ~2,000 cells) or E. coli (red bars, ~10,000) cells classified by 16S rRNA sequencing. The numbers on the bars represent the mean number of cells detected. Data points represent independent replicates. O/N, overnight cultures. f, A mock community is generated by combining cultures of eight different bacteria in specific proportions. Relative abundance of the community is determined by DoTA-seq and plotted next to the theoretical values. C. hiranosis, Clostridium hiranonis; L. lactis, Lactococcus lactis.