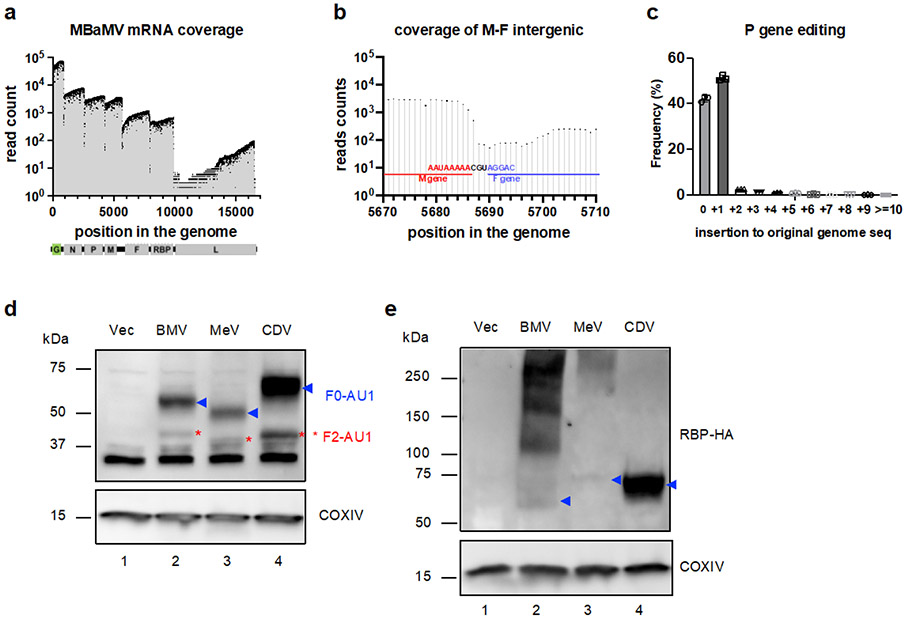
Extended Data Figure 5. Molecular characterization of MBaMV transcripts.
Vero-bCD150 cells were infected with MBaMV (MOI=0.01) and cytosolic RNA collected at 2 dpi were subjected to direct RNA sequencing targeting mRNAs using the MinION platform (Oxford Nanopore Technology). 93,917 reads / total 797,133 reads are aligned to MBaMV. a, Read coverage showed that MbaMV infection and replication exhibited the 3’ to 5’ transcriptional gradient typical of paramyxoviruses. The boundary of each transcriptional unit can also be appreciated by the steep drop in reads that coincide with the tri-nucleotide intergenic sequence. Median (IQR) sequencing coverage ranges are 6509 (N, 5368-7465), 3640 (P, 3193-4047), 3018 (M, 2533-3656), 929 (F, 722-1096), 535 (RBP, 491-633) and 10 (L, 5-33). b, The read coverage graph around the M-F intergenic region is magnified to show the uncommon intergenic motif of ‘CGU’. The intergenic motif between all other genes is ‘CUU’. c, Amplicon sequencing around the P gene editing site shows the frequency of P gene-mRNA editing. 51.2% of P gene mRNAs acquired a single ‘G’ insertion at the editing site, creating V mRNA. The numbers shown are the average of three independent experiment with error bars representing S.D. (d-e) show the expression of surface glycoproteins (RBP and F) of MBaMV, MeV, and CDV by Western blot. 5 μg expression plasmids (pCAGGS, pCAGGS-RBP-HA, pCAGGS-F-AU1) were transfected into 293T in 6-well plate. Cytosolic proteins were collected and run on acrylamide gel. AU1 staining (d) showed full-length F protein (F0; blue arrowhead) and cleaved F1 (red asterisk) product. HA staining (e) showed expression of RBP monomer (blue arrowhead) around 60 – 70 kDa in addition to oligomer. COXIV was used as loading control for blot. The experiments in d) and e) were completed twice, with similar results.