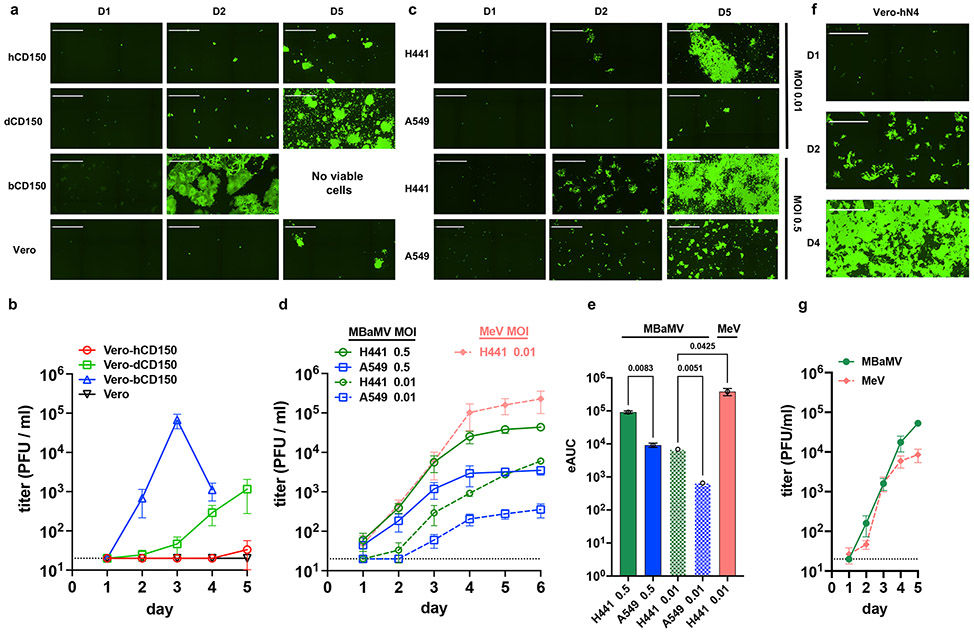
Figure 3. MBaMV replicates efficiently in cells expressing bCD150 and human nectin-4.
a-b, Vero-hCD150, Vero-dCD150, Vero-bCD150, and Vero cells were infected with rMBaMV-EGFP (MOI 0.01). Virus replication and spread were monitored by imaging cytometry (a) and virus titer in the supernatant (b). a, Large syncytia were evident in Vero-bCD150 cells by 2 dpi. b, Supernatant was collected every day and the virus titer was determined by a GFP plaque assay (see methods). Data shown are mean +/− S.D. from triplicate experiments. c-e, H441 and A549 cells were infected with rMBaMV-EGFP at a low (0.01) or high (0.5) MOI. Virus replication and spread were monitored as in a-b. c, Infected H441 and A549 cells at 1, 2 and 5 dpi (D1, D2, D5). d, Virus growth curves represented by daily titers in the indicated conditions. Data shown are mean titers +/− S.D. from triplicate infections. e, The empirical Area Under Curve (eAUC) was obtained from each growth curve and plotted as a bar graph from 3 independent experiments (mean +/− S.D.) (PRISM v 9.0). The dotted bars represent the growth curves done at an MOI of 0.1 and the filled bars at MOI 0.5. Adjusted p values are indicated (one-way ANOVA Dunnett’s T3 multiple comparison test). f-g, Vero-human nectin-4 cells (Vero-N4) were infected with MBaMV and MeV (MOI 0.01). f, MBaMV infected Vero-hN4 at D1, D2 and D4. g, Replicative virus titers for MBaMV and MeV on Vero-hN4 cells over 5 days (mean +/− S.D., n=3). White bar in a, c, and f equals 1 millimeter. All images shown are captured by a Celigo Imaging Cytometer (Nexcelom). Images are computational composites from an identical number of fields in each well. The limit of detection for virus titer determination is 20 PFU/ml and is indicated by the dotted line in b, d, and g.