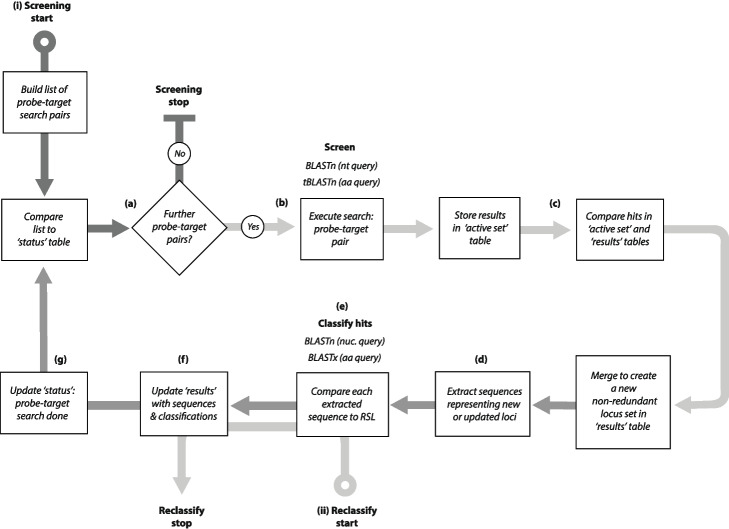
Fig. 2.
The database-integrated genome screening (DIGS) process as implemented in the DIGS tool. (i) Screening. a On initiation of screening a list of searches, composed of each query sequence versus each target database (TDb) file is composed based on the probe and TDb paths supplied to the DIGS program. Subsequently, screening proceeds systematically as follows: b the status table of the project-associated screening database is queried to determine which searches have yet to be performed. if there are no outstanding searches then screening ends, otherwise it proceeds to step b wherein the next outstanding search of the TDb is performed using the selected probe and the appropriate BLAST+ program. Results are recorded in the data processing table (“active set”); c results in the processing table are compared to those (if any) obtained previously to derive a non-redundant set of non-overlapping loci, and an updated set of non-redundant hits is created, with each hit being represented by a single results table row. To create this non-redundant set, hits that overlap, or occur within a given range of one another, are merged to create a single entry. d Nucleotide sequences associated with results table rows are extracted from TDb files and stored in the results table; e extracted sequences are classified via BLAST-based comparison to the RSL using the appropriate BLAST program. f The header-encoded details of the best-matching sequence (species name, gene name) are recorded in the results table. g The status table is updated to create a record of the search having been performed, and the next round of screening is initiated. (ii) Reclassification: hits in the results table can be reclassified following an update to the reference sequence library