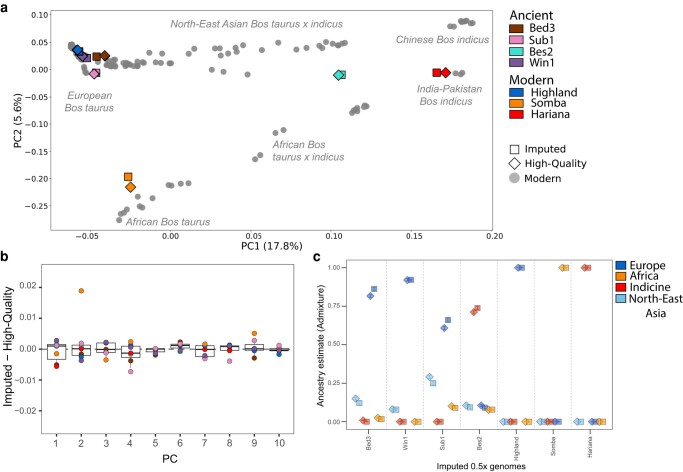
Fig. 3.
PCA and ADMIXTURE of imputed and high-quality genotypes of the seven test samples onto the modern cattle reference panel. a) Projections for 0.5× imputed and high-quality diploid genotypes of the test samples onto the modern reference panel along the first two eigenvectors. Data set was filtered for a MAF ≥ 2.5%, transversions, and LD pruning resulting in 812,801 SNPs. Test samples are denoted by color with imputed and high-quality represented by squares and diamonds, respectively, while the reference panel individuals are plotted as circles. b) Boxplots of the normalized differences in the coordinates of the high-quality and 0.5× imputed genomes for the first ten principle components. The horizontal lines of the boxplot represent the first quartile, median, and third quartile; whiskers represent 1.5 times the quartile range. c) Comparison of the ancestral components modelled by ADMIXTURE (K = 4) based on 800,694 SNPs (MAF ≥ 2.5%, transversions and LD pruning) run with the reference panel and the seven test samples. Here we display results for the 0.5x imputed (squares) and high-quality validation (diamonds) genotypes. Error bars represent the deviation from the mean measured as standard errors calculated from 1,000 replicates.