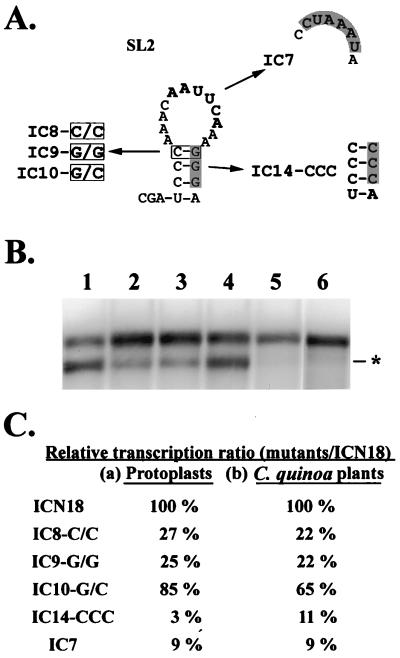
FIG. 4.
Predicted secondary structure of SL2 and the schematic diagrams of satBaMV mutants (A) and their transcription activities (B and C). (A) Shown are the predicted secondary structure of SL2 and the schematic diagrams of mutants introduced in SL2. The boxed and shaded letters represent the mutated nucleotides. The bold letters indicate the conserved nucleotide motif. (B) Northern blot analysis of total RNAs extracted from 5 × 104 protoplasts of N. benthamiana coinoculated with BaMV-L RNA and pICN18 (lane 1), pIC8-C/C (lane 2), pIC9-G/G (lane 3), pIC10-G/C (lane 4), pIC14-CCC (lane 5), and pIC7 (lane 6) transcripts at 24 h.p.i. with a satBaMV-specific riboprobe. The asterisk indicates the position of the 0.7-kb sgRNA transcribed from the satBaMV cassette. (C) The relative ratio was the transcription ratio of a mutant over that of pICN18 in protoplasts of N. benthamiana (a) or in leaves of C. quinoa (b). These data were collected three times from protoplasts and two times from plants.