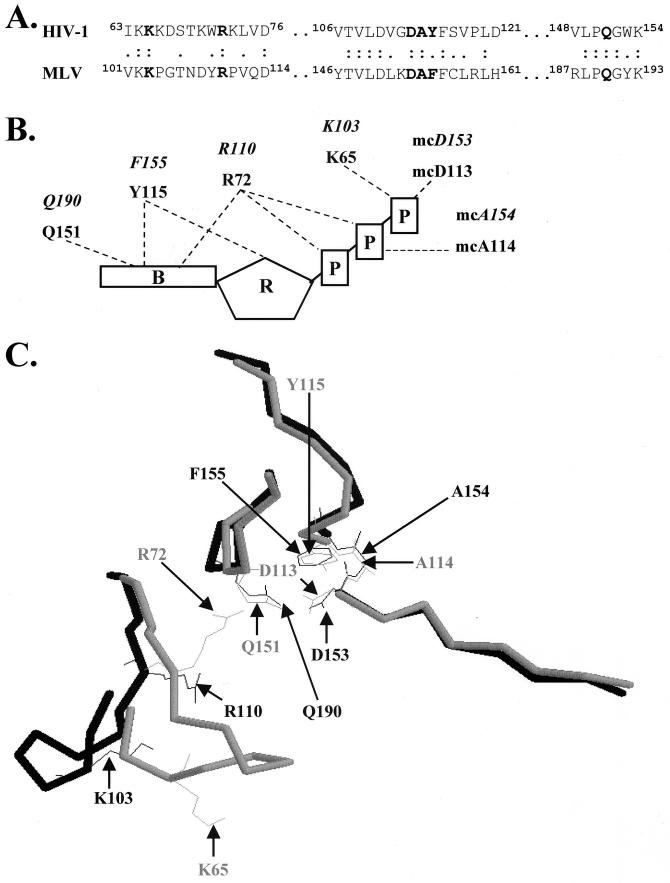
FIG. 2.
The dNTP-binding sites of MLV and HIV-1 RTs. (A) Primary sequence homology of the MLV and HIV-1 RT dNTP-binding sites. HIV-1 and MLV RTs were aligned through their TVLD, LPQG, and YXDD amino acid motifs. Residues of the dNTP-binding site of MLV RT were determined by further alignment with residues proposed to constitute the dNTP-binding site of HIV-1 RT. Two dots represent residues that are identical, whereas one dot represents conservative changes between the two RTs. Boldface lettering represents residues constituting the dNTP-binding sites in both HIV-1 and MLV RTs. (B) Structure of HIV-1 RT dNTP-binding site (modified from Huang et al. [20]). The structure shown contains amino acid positions K65, R72, D113, A114, Y115, and Q151 interacting with the dTTP substrate. Italic lettering represents the equivalent residues in MLV RT constituting the dNTP-binding site. Components of the dTTP substrate are denoted as B (base), R (ribose sugar), and P (phosphate). Dashed lines represent interactions between the amino acid residues of RT and the dTTP substrate. mc, main chain interactions between the substrate and the RT. (C) Comparison of structures of MLV and HIV-1 RTs constituting the dNTP-binding sites (13, 19). The backbone of HIV-1 RT (amino acid sequences 63 to 76, 106 to 121, and 148 to 154) was superimposed on the homologous structure in MLV RT (amino acid sequences 101 to 114, 146 to 161, and 187 to 193). Free HIV-1 RT (not bound to DNA or substrate dNTP) is represented in gray; MLV RT is represented in black. Side chains of residues creating the dNTP-binding sites of MLV and HIV-1 RTs are illustrated as wire frames. The structures were superimposed using the RasMol Program to maximize the overlaps between the peptide backbones of the HIV-1 and MLV RTs.