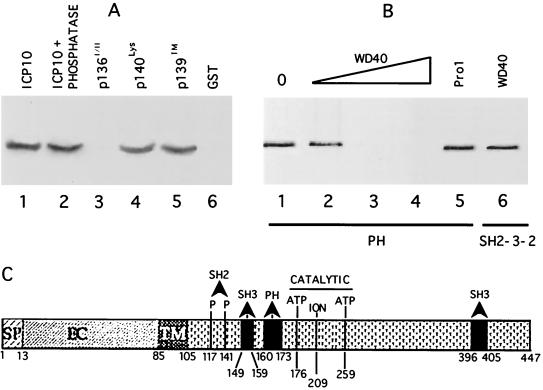
FIG. 5.
Ras-GAP PH binds a WD-40 like site in ICP10 PK. (A) In vitro binding assays with Ras-GAP fusion protein PH (lanes 1 to 5) or uncoated beads (lane 6) and extracts of cells that constitutively express the autophosphorylated ICP10 untreated (lanes 1 and 6) or treated (2 h, 37°C) with 15 U of CIAP (lane 2) or extracts of cells that constitutively express ICP10 mutants p136I/II (lane 3), p140lys (lane 4), or p139TM (lane 5). Uncoated beads are shown in lane 6. (B) Peptide competition assays using beads coated with Ras-GAP fusion protein PH (lanes 1 to 5) or SH2-SH3-SH2 (lane 6), extracts of cells that constitutively express ICP10 (lane 1), and peptide WD40 at 10 μM (lane 2), 50 μM (lane 3), or 100 μM (lanes 4 and 6) or peptide Pro1 (100 μM; lane 5). (C) Schematic model of known sites in ICP10 PK includes signal peptide (SP; residues 1 to 13), extracellular domain (EC), transmembrane domain (TM; residues 85 to 105), binding sites for RAS-GAP N-SH2 (residues pThr117 and pThr141), and PH (residues 160 to 173) domains, Grb2 SH3 binding site (residues 149 to 159 and 396 to 405), and catalytic core (ATP-binding Lys176 and Lys259 and ion-binding Glu209).