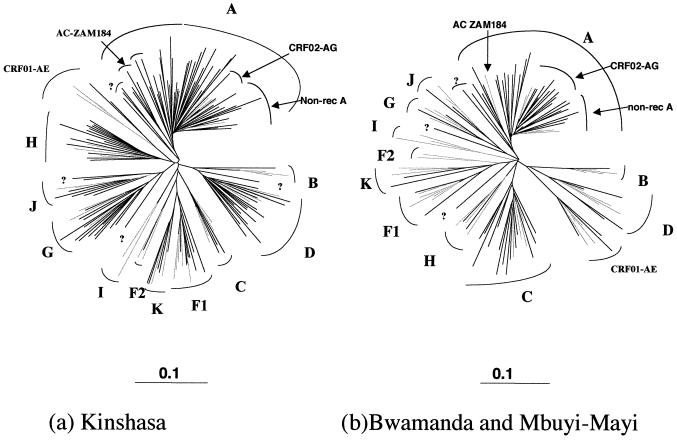
FIG. 2.
Phylogenetic tree based on 441 unambiguously aligned nucleotides from the env V3-V5 region of the 197 new HIV-1 isolates, from Kinshasa (a) and from Bwamanda and Mbuyi-Mayi (b), and reference strains representing the different genetic subtypes: A-KE.Q2317, A-SE.SOSE7253, A-92UG037, CRF02-AG-IBNG, CRF02-AG-DJ263, CRF02-AG-DJ264, B-RF, B-WEAU160, B-JRFL, B-HBX2, C-ETH2220, C-92BR025, C-IN.21068, C-BW.96BW0502, D-NDK, D-ZR.84ZR085, D-94UG114, D-ELI, CRF01-AE-90CR402, CRF01-AE-93TH253, CRF01-AE-CM240, F1-93BR020, F1-BZ163, F1-BZ126, F1-BE.VI850, F1-FI.FIN6393, F2-95CMMP255, F2-95CMMP257, G-BE.DRCBL, G-HH8793, G-SE6165, H-90CF056, H-BE.VI991, H-BE.VI997, J-SE.SE91733, J-SE.SE92809, K-96CMMP535, and K-97ZREQTB11. The analysis was performed as described in Materials and Methods. Reference strains are in grey, and strains from the DRC are indicated in black. Non-rec, nonrecombinant.