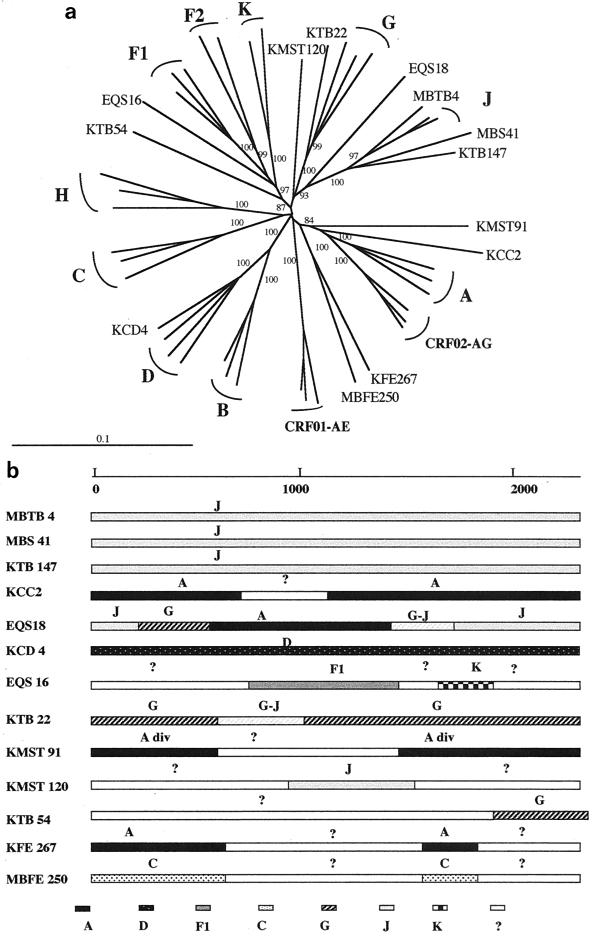
FIG. 5.
(a) Unrooted phylogenetic tree based on 2,278 unambiguously aligned nucleotides from the entire gp160 gene from 13 new HIV-1 isolates and reference strains representing different genetic subtypes. Analysis was performed as described in Materials and Methods; the reference strains used in the phylogenetic tree analysis were A-KE.Q2317, A-SE.SOSE7253, A-92UG037, CRF02-AG-IBNG, CRF02-AG-DJ263, CRF02-AG-DJ264, B-WEAU160, B-JRFL, B-HBX2, C-ETH2220, C-IN.21068, C-BW.96BW0502, D-NDK, D-ZR.84ZR085, D-94UG114, CRF01-AE-90CR402, CRF01-AE-93TH253, CRF01-AE-CM240, F1-93BR020, F1-BE.VI850, F1-FI.FIN6393, F2-95CMMP255, F2-95CMMP257, G-BE.DRCBL, G-HH8793, G-SE6165, H-90CF056, H-BE.VI991, H-BE.VI997, J-SE.SE91733, J-SE.SE92809, K-96CMMP535, and K-97ZREQTB11. (b) Putative recombination breakpoints within the gp160 gene were localized based on DIVERT and bootscan analysis (see Materials and Methods) using the reference strains listed above. The results were confirmed by phylogenetic tree analysis with 1,000 bootstrap replicates. Subtypes were designated when the bootstrap values were above 800, divergent subtypes were designated when bootstrap values were between 650 and 800, and fragments were identified as unclassified when they did not cluster at all with any of the known subtypes.