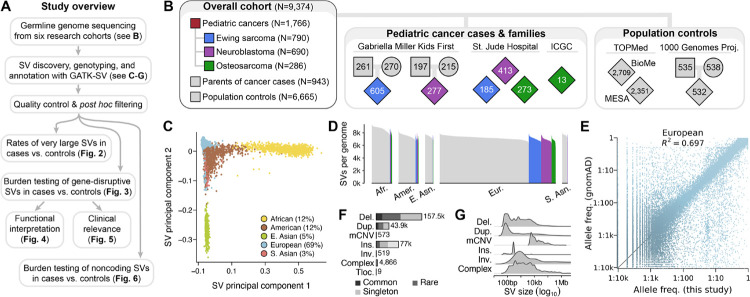
Fig. 1 |. The landscape of germline SVs in 9,374 pediatric cancer patients and controls.
(A) Roadmap of key datasets and analyses in this study. (B) Summary of study cohort structure after excluding low-quality WGS samples. (C) A principal component analysis of common, high-quality SVs stratified individuals based on their genetic ancestry. See also Fig. S2. (D) We detected an average of 7,276 high-quality SVs per genome, which was correlated with sample ancestry as expected based on human demographic history.18 (E) The allele frequencies (AFs) of SVs detected in this study were strongly correlated with AFs of SVs reported from WGS of 63,046 unrelated individuals in the Genome Aggregation Database (gnomAD).17 European samples shown here; see Fig. S3 for other ancestries. (F-G) Most SVs were small (<1kb) and rare (AF<1%). Del.: deletion. Dup.: duplication. mCNV: multiallelic CNV. Ins.: insertion. Tloc.: reciprocal translocation.