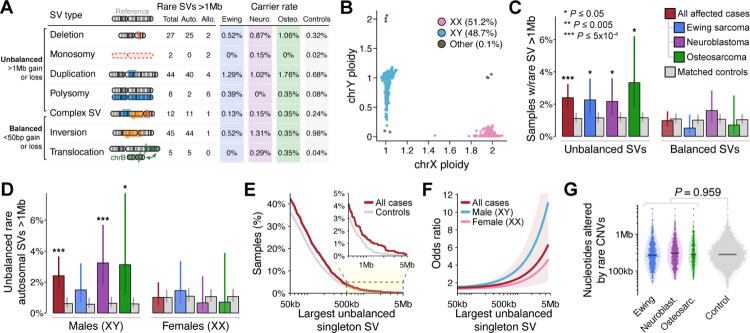
Fig. 2 |. Very large germline CNVs increase risk for pediatric solid tumors in a sex-specific manner.
(A) We identified 143 rare (AF<1%), large (>1Mb) germline SVs in a set of 1,745 pediatric cancer cases and 4,983 ancestry-matched adult controls. Auto.: autosomal. Allo.: allosomal. Neuro.: neuroblastoma. Osteo.: osteosarcoma. (B) WGS delineated sample sex based on ploidy (i.e., copy number) estimates of X and Y chromosomes. (C) We discovered an association between pediatric solid tumors and the 84 unbalanced SVs from (A). We did not observe any significant associations with comparably large but balanced rare SVs. Error bars indicate 95% confidence intervals (CI); P values derived from logistic regression adjusted for sex, cohort, and ancestry. (D) Results from (C) restricted to autosomes and stratified by male (karyotypic XY) vs. female (karyotypic XX) samples. (E) Proportion of cases and controls who carried at least one singleton unbalanced germline SV larger than the size specified on the X-axis. See also Fig. S4F. (F) Relationship between the size of unbalanced singleton SV size and the corresponding odds ratio for pediatric cancer based on the data from (D) presented as a cubic smoothing spline. Shaded area is 95% CI for a pooled model of both sexes from all histologies. (G) The total sum of nucleotides altered by autosomal, rare, unbalanced SVs did not significantly differ between cases & controls.