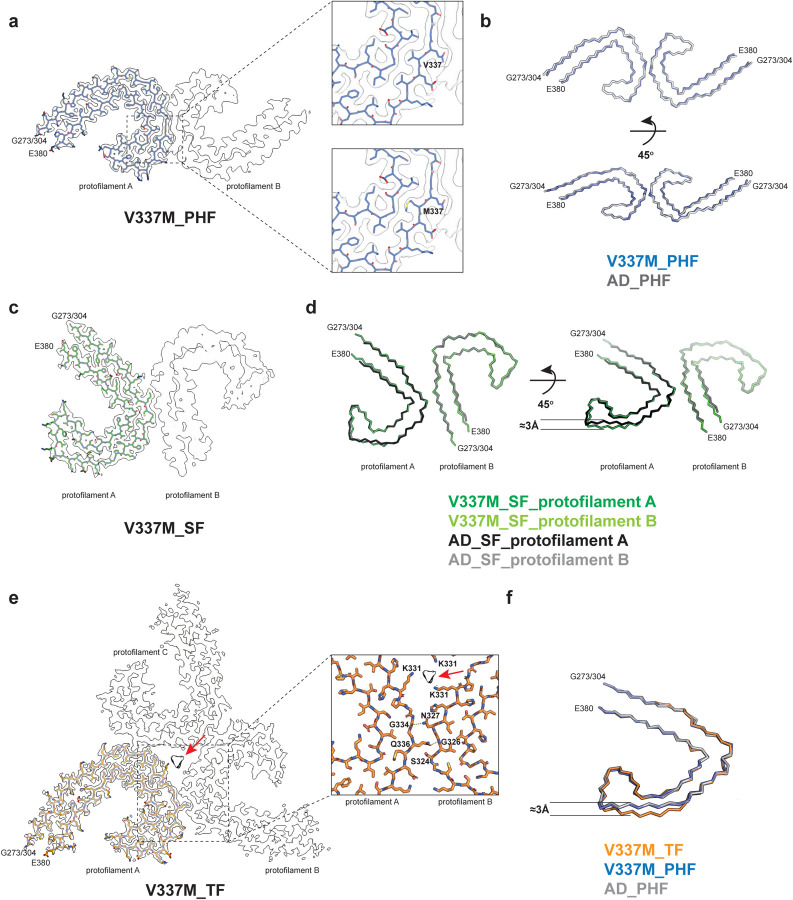
Figure 2. V337M mutation in MAPT: Cryo-EM structures of tau filaments.
a, Cryo-EM density map and atomic model of paired helical filament (PHF). Two identical protofilaments extend from G273/304-E380. Inset: Zoomed-in view showing that both wild-type (V) and mutant (M) residues can fit into the density at position 337.
b, Backbone representation of overlay of PHF extracted from the frontal cortex of case 1 with mutation V337M in MAPT (blue) and PHF extracted from the frontal cortex of an individual with sporadic AD (white, PDB:5O3L). The root mean square deviation (rmsd) between Cα atoms of the two structures is 0.78 Å.
c, Cryo-EM density map and atomic model of straight filament (SF). Two asymmetrically packed protofilaments A and B extend from G273/304–380.
d, Overlay of SF extracted from the frontal cortex of case 1 with mutation V337M (protofilament A is in dark green and protofilament B is in light green) and SF extracted from the frontal cortex of an individual with AD (PDB:5O3T) (protofilament A is in black and protofilament B is in grey). In protofilament A, strand β4 (residues 336–341) is shifted along the helical axis by 3 Å. Protofilament B adopts the same structure as in AD. e, Cryo-EM density map and atomic model of triple filament (TF). Three identical protofilaments (A, B and C) extend from G273/304-E380. An additional non-proteinaceous density at the filament’s three-fold axis is labelled with a red arrow. Inset: Zoomed-in view showing one of the three identical protofilament interfaces and K331 residues from each protofilament coordinating the additional density.
f, Overlay of individual protofilaments from TF with mutation V337M (orange), PHF with mutation V337M (blue) and PHF from AD (white), viewed at a 45° angle to the filaments’ axes, as in panel d.