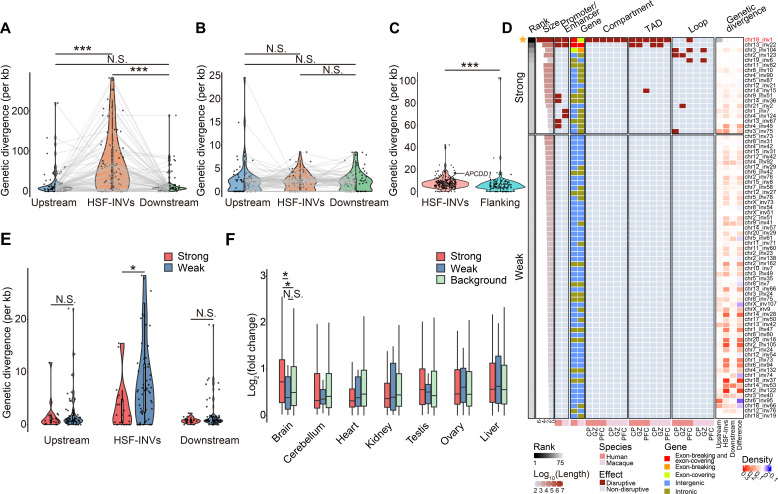
Fig. 6. Characteristics of human-specific inversions.
(A) Violin plots showing the genetic divergences of fixed human-specific inversions (HSF-INVs) and their length-matched, upstream and downstream genomic regions (Upstream and Downstream regions). Wilcoxon signed rank tests were performed. Wilcoxon signed rank tests, ***P < 0.001. (B) Violin plots showing the genetic divergences of homologous macaque regions of the HSF-INVs and their upstream and downstream regions. Wilcoxon signed rank tests; N.S., not significant. (C) Violin plots showing the genetic divergences of promoter regions in fixed human-specific inversions (HSF-INVs) and promoter regions in length-matched flanking genomic regions (Flanking regions). The red dot indicates promoter of APCDD1. Wilcoxon rank-sum test, ***P < 0.001. (D) Classification of 75 fixed human-specific inversions into two groups with different degree of regulatory effects (Strong and Weak), based on their sizes and locations in the human and macaque genomes. The genetic divergence relative to the human-chimpanzee common ancestor is also shown for the 75 HSF-INVs and corresponding upstream and downstream regions. The difference in the genetic divergence between each inversion and the average of its upstream and downstream regions is also shown (Difference). (E) Violin plots showing the genetic divergence for inversions with strong (Strong) or weak (Weak) effects, and their upstream and downstream regions. One-sided, Wilcoxon rank-sum tests, *P < 0.05, N.S., not significant. (F) The log2-transformed fold changes in gene expression in the fetal brain between humans and macaques, for genes located on inversions with strong (Strong) or weak (Weak) effects, as well as for genome-wide orthologs as a background (Background). Wilcoxon rank-sum tests, *P < 0.05, N.S., not significant.