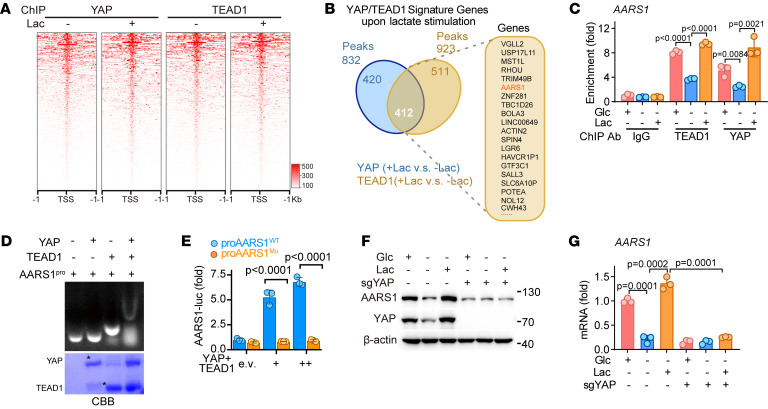
Figure 4. AARS1 and YAP-TEAD form a positive-feedback loop.
(A) ChIP-Seq analysis heatmap representing the distribution of YAP or TEAD1 binding relative to the gene transcription start site (TSS) in cells cultured in glucose-free medium treated with or without lactate. Lac, lactate. (B) Venn diagram illustrating the overlap of YAP- and TEAD1-enriched genes upon lactate stimulation. The top 20 genes are shown. (C) ChIP-qPCR showing the enrichment of YAP and TEAD1 on the AARS1 promoter in lactate-treated HGC27 cells (n = 3). Glc, glucose. Data are presented as mean ± SD. (D) Top: Gel shift analysis showing the binding of YAP and TEAD1 to the synthetic DNA probe containing TEAD1-binding site on the AARS1 promoter. Bottom: CBB staining of purified recombinant YAP (MBP-YAP 1-291) and TEAD1 proteins used in gel shift assay. (E) Luciferase activity of WT or mutant (Mu) AARS1 promoter vectors in YAP- and TEAD1-overexpressing HEK293FT cells (n = 3). Data are presented as mean ± SD. (F) Immunoblotting of AARS1 protein levels in YAP-knockout AGS cells upon lactate treatment. (G) AARS1 mRNA levels in YAP-knockout cells upon lactate stimulation (n = 3). Data are presented as mean ± SD. Unpaired 2-tailed Student’s t test (C, E, and G).