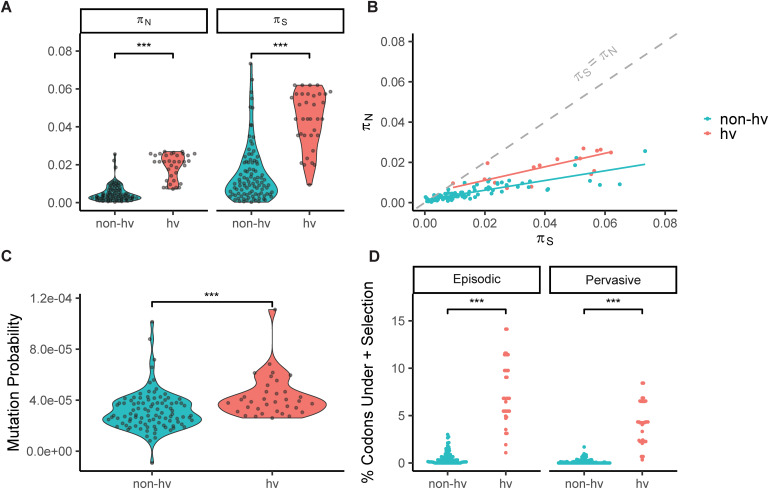
Figure 6. hvNLR nucleotide diversity is associated with a high likelihood of mutation and codons under diversifying selection.
(A) Average nonsynonymous pairwise nucleotide diversity per site (πN) and average synonymous pairwise nucleotide diversity per site (πS). (B) πS vs πN of the coding sequence of NLRs with per group linear regressions. The neutral expectation, πS = πN, is shown as a gray dotted line. (C) Mutation probability score of hv and non-hvNLRs. (D) Percentage of codons under positive selection determined by MEME (episodic), and FEL (pervasive). Data Information: For all comparisons, n = 97 for non-hvNLRs and n = 35 for hvNLRs, with n referring to the number of genes tested. The significance shown is the result of unpaired Wilcoxon rank-sum tests, with *** indicating a P value < 0.001. Source data are available online for this figure.