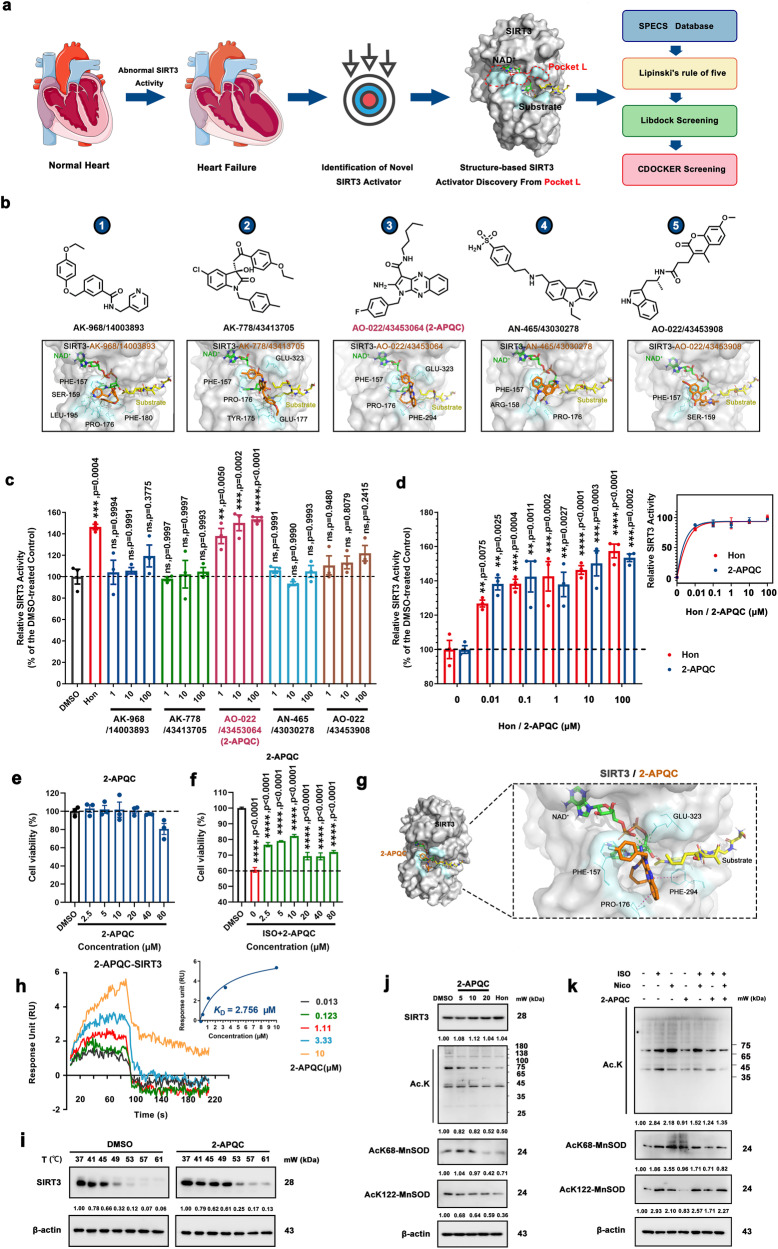
Fig. 1.
Structure-based screening of small-molecule activators of SIRT3 and identifies 2-APQC activates SIRT3 deacetylase activity in H9c2 cells. a The structure of SIRT3 and virtual screening scheme model for the discovery of SIRT3 activator. Created with biorender.com and https://smart.servier.com. b The 5 candidate compounds based on virtual screening and molecular docking. c The relative SIRT3 deacetylation activity after the candidate compounds (1, 10, 100 μM) and Honokiol (Hon) (10 μM) treatment. Data are present as mean ± s.e.m, n = 3. d The relative SIRT3 deacetylation activity after Hon or 2-APQC (0.01, 0.1 1, 10, 100 μM) treatment. Data are present as mean ± s.e.m, n = 3. e H9c2 cells were treated with indicated concentration of 2-APQC for 24 h, and cell viability was measured by the MTT assay. Data are present as mean ± s.e.m, n = 3. f H9c2 cells were treated with indicated concentration of 2-APQC for 24 h, and then treated with isoproterenol (ISO) for 48 h, the cell viability was measured by the MTT assay. Data are present as mean ± s.e.m, n = 3. g The molecular docking sketch map to indicate the binding mode between SIRT3 and 2-APQC. h Surface plasmon resonance assay detected the Kd value of SIRT3 in H9c2 cells treated with 2-APQC. i Cellular thermal shift assay detected the thermal stability of SIRT3 in H9c2 cells treated with 2-APQC. j Western blot analysis of SIRT3, acetylated-lysine, acetylated manganese superoxide dismutase 2 (MnSOD2) at K68 and K122 protein expression in H9c2 cells treated with 2-APQC for 24 h. k Western blot analysis of acetylated-lysine, acetylated MnSOD2 at K68 and K122 protein expression in H9c2 cells treated with 2-APQC for 24 h, together with or without ISO/nicotinamide (Nico) as the research design. β-actin, loading control. ns no significance; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Hon Honokiol, Nico nicotinamide, ISO isoproterenol