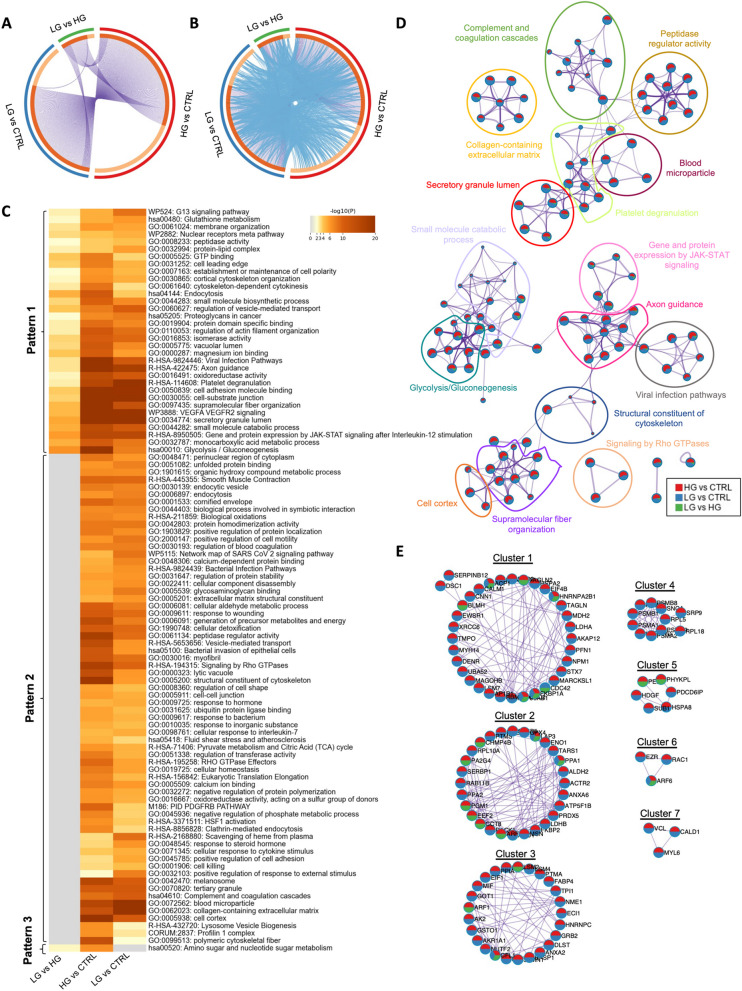
Fig. 4.
Visualisation of the functional enrichment meta-analysis based on three protein lists [LG vs. control (CTRL), HG vs. control (CTRL) and LG vs. HG)] in soft mucin samples compared to control tissues. A Circos plot visualising the overlap between the protein lists. Each candidate protein is assigned to a point on the arc of the corresponding protein list(s). Proteins common to both lists are connected by purple curves. B Circos plot visualisation with blue curves connecting those candidate proteins that have different identities but share an enriched pathway/process, i.e. they represent the functional overlap between protein lists. C Heatmap showing the top 100 enrichment clusters, one row per cluster, using a discrete colour scale to represent statistical significance. Grey colour indicates the lack of enrichment for that term in the corresponding gene list, light yellow colour indicates the boundary between significance and insignificance, deep yellow colour indicates a high degree of significance. D Enrichment network visualisation for results from the three protein lists, where nodes are represented by pie charts indicating their associations with each input list. Cluster labels have been added manually. Colour code represents the identities of protein lists, where blue indicates LG vs. CTRL, red indicates HG vs. CTRL and green indicates LG vs. HG. E Visualisation of the PPI network and MCODE components identified from the combined protein list, where each node represents a protein with a pie chart encoding its origin. Colour codes for pie sectors represent a protein list