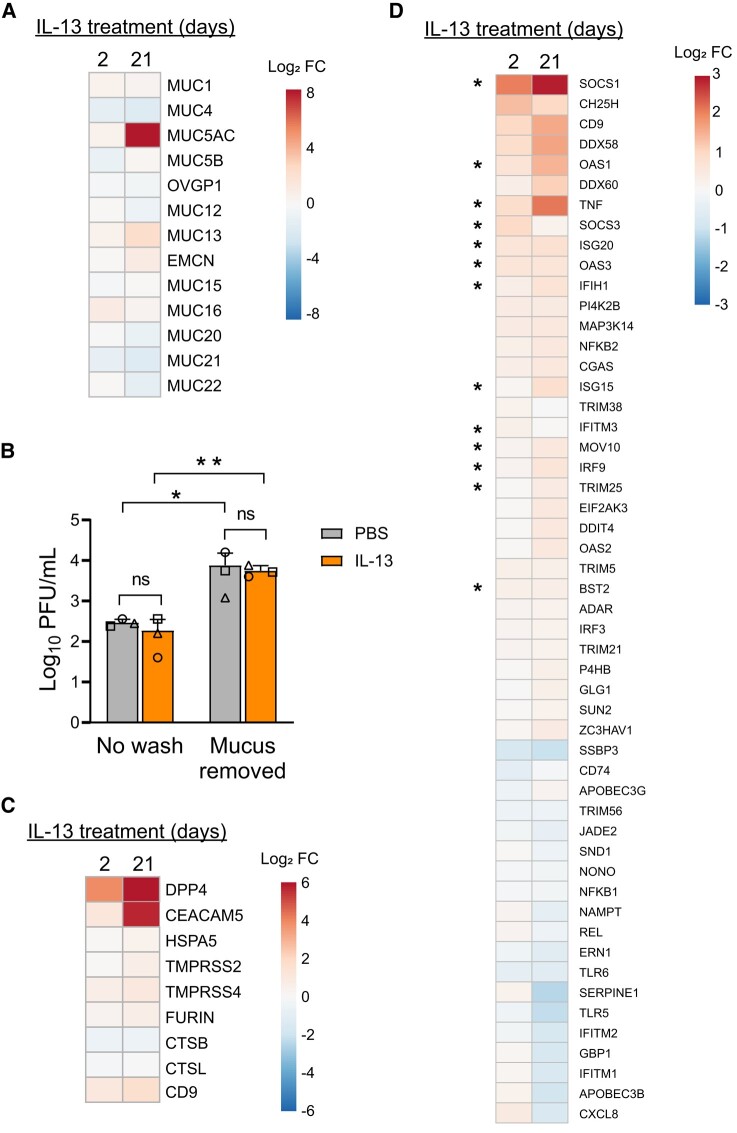
Figure 5.
IL-13 treatment alters expression of genes that influence MERS-CoV entry and infection, antiviral defense, and the mucin barrier. A, HAE (n = 8) were treated with PBS or IL-13 (20 ng/mL) for 2 or 21 days, and transcriptional profiling was carried out by bulk RNAseq. Displayed on the heat map are expression changes in all detected mucin genes after 2 or 21 days of IL-13 treatment. Expression levels are represented as log2 fold change. B, HAE (n = 3) were stimulated with PBS or IL-13 (20 ng/mL) for 21 days. Accumulated mucus on the apical surface was removed by rinsing with 10mM DTT (“Mucus removed”), while control cells were not rinsed (“No wash”). Epithelia were then infected with MERS-CoV (MOI, 0.5), and apical surface liquid was collected at 1 day postinfection for titering. Data are presented as mean ± SD. Log-transformed data were tested for statistically significant differences between conditions by 2-way analysis of variance, followed by a Sidak multiple-comparison test. *P < .05. **P < .01. C, Heat map depicts genes with known roles in MERS-CoV attachment/entry that were differentially expressed in IL-13–treated epithelia relative to PBS-treated cells (P < .01 for at least 1 treatment condition). D, Heat map summarizes interferon-stimulated genes that were up- or downregulated by IL-13 treatment in the RNAseq data set (P < .01 in at least 1 treatment condition). Asterisks denote genes that have been reported to be targets of STAT6 signaling. All heat maps were generated through ClustVis (https://biit.cs.ut.ee/clustvis/). DTT, dithiothreitol; FC, fold change; HAE, human airway epithelia; IL-13, interleukin 13; MOI, multiplicity of infection; ns, not significant; PBS, phosphate-buffered saline; PFU, plaque-forming units; RNAseq, RNA sequencing.