Fig. 1.
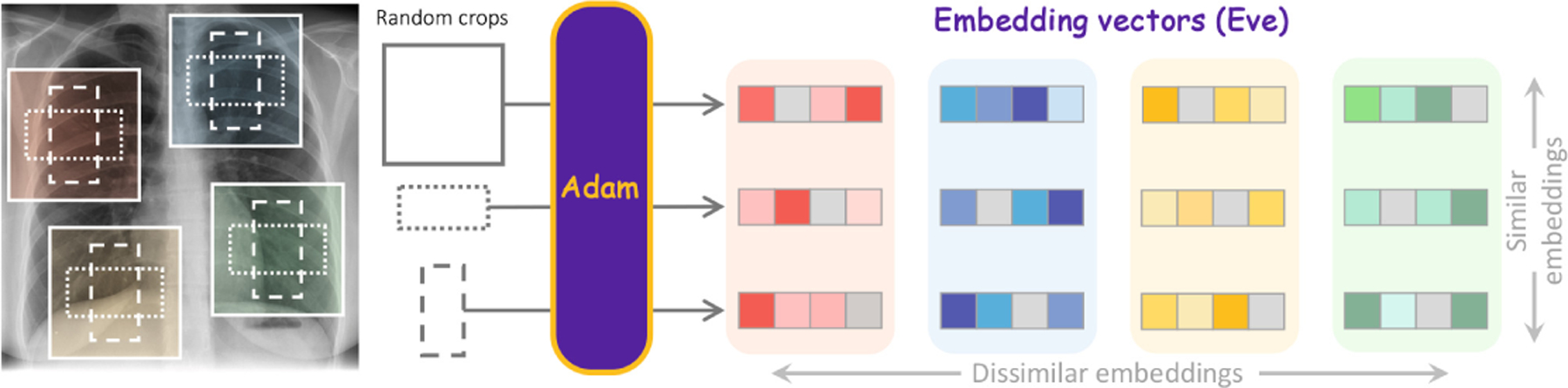
Existing SSL methods lack capabilities of “understanding” the foundation of medical imaging—human anatomy. We believe that a foundation model must be able to transform each pixel in an image (e.g., a chest X-ray) into semantics-rich numerical vectors, called embeddings, where different anatomical structures (indicated by different colored boxes) are associated with different embeddings, and the same anatomical structures have (nearly) identical embeddings at all resolutions and scales (indicated by different box shapes) across patients. Inspired by the hierarchical nature of human anatomy (Fig. 6 in Appendix), we introduce a novel SSL strategy to learn anatomy from medical images (Fig. 2), resulting in embeddings (Eve), generated by our pretrained model (Adam), with such desired properties (Fig. 4 and Fig. 8 in Appendix).
