Figure 3.
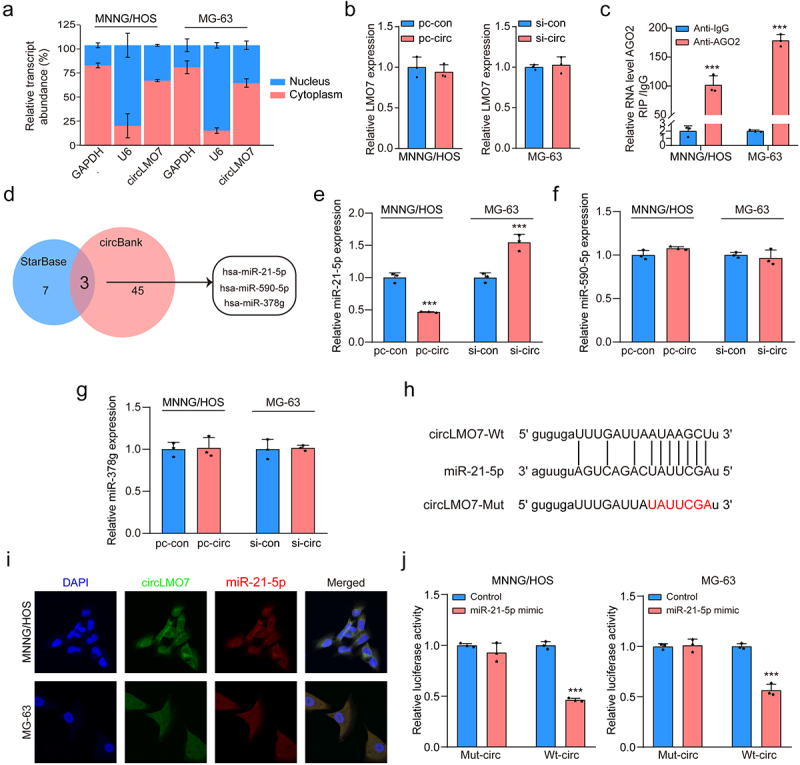
Circ-LMO7 directly targets on miR-21-5p in OS cells.
(a, b) Subcellular localization of circ-TRIM35 in MNNG/HOS and MG-63 cells, as determined using PCR. (b) qRT – PCR analysis of LMO7 expression in transfected MNNG/HOS and MG-63 cells (c) The enrichment of circ-LMO7 in the immunoprecipitation products was detected by qPCR. (d) The miRNAs that may bind with circ-LMO7 were predicted through StarBase database and CircBank database. (e, f, & g) MiR-21-5p, miR-590-5p and miR-378 g expression in transfected MNNG/HOS and MG-63 cells was examined by qRT-PCR. (h) MiR‐21-5p binding sequences in circ-LMO7 are displayed, and reporter gene plasmids were constructed. (i) Dual RNA-FISH and immunofluorescence staining assays indicating the co-localization of circ-LMO7 (red) and miR-21-5p (green) in MNNG/HOS and MG-63 cells, with nuclei staining using DAPI (blue). (j) The regulatory relationships were confirmed by luciferase reporter assay. *p < .05, **p < .01, ***p < .001
