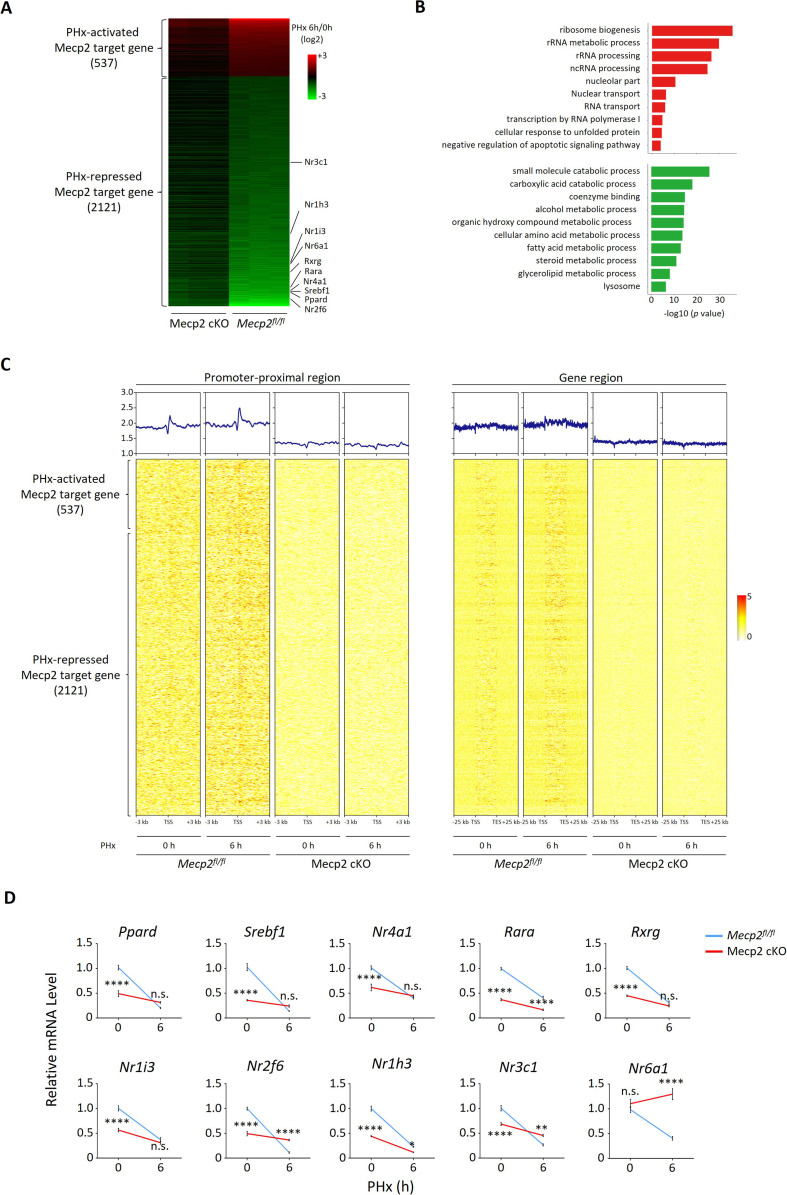
Figure 6. Mecp2 transcriptionally regulates quiescence exit.
(A) Heatmap of Mecp2 direct target genes at the early stage of liver regeneration rank-ordered by their gene expression fold change. (B) The top 10 most significantly overrepresented Gene Ontology (GO) terms for the partial hepatectomy (PHx)-activated (red) and PHx-repressed (green) Mecp2 target genes. (C) Heatmaps depicting ChIP-seq enrichment of Mecp2 at the promoter-proximal region (3 kb away from transcription start sites [TSS]) and the defined gene region of Mecp2 target genes in Mecpfl/fl and Mecp2 cKO livers before and 6 hr post-PHx. Genes are rank-ordered according to the fold change of expression. (D) Real-time PCR validation of PHx-repressed NRs in Mecp2fl/fl and Mecp2 cKO livers upon PHx. Data are presented as means ± SEM; n = 5. n.s., not significant; **p<0.01; ****p<0.0001 by two-way ANOVA.