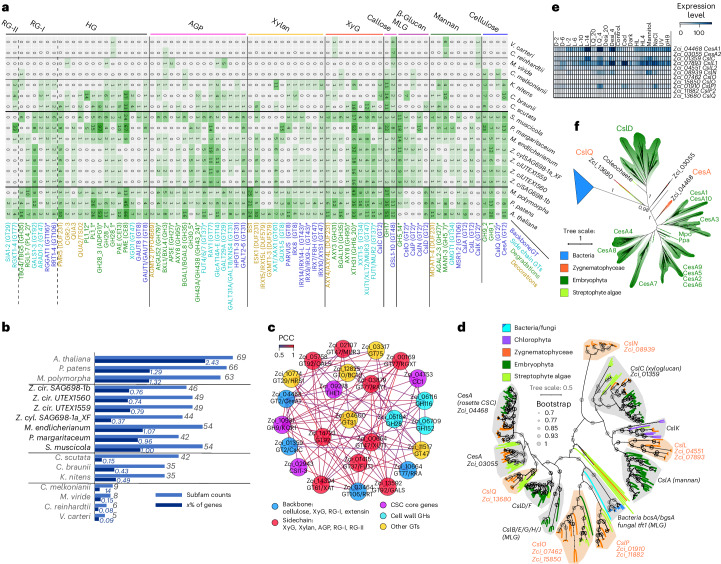
Fig. 4. Cell wall innovations revealed by protein family analyses.
a, A heatmap of homolog presence in 77 enzyme subfamilies (rows) across 17 plant and algal genomes (except for Coleochaete scutata, for which we used its transcriptome). The enzyme subfamilies are grouped by polysaccharide, and the colors indicate their biochemical roles; phylogenetic patterns compatible with gene gains that might have involved HGT are indicated with asterisks. b, Counts of subfamilies and gene percentages (with respect to the total annotated genes) across the 17 species. Shown in the plot is the gene percentage × 100. c, The co-expression network of SAG 698-1b containing 25 genes (most belonging to the 77 analyzed subfamilies) involved in cell wall polysaccharide syntheses. d, The phylogeny of GT2 across the 17 species. Major plant CesA/Csl subfamilies are labeled by the SAG 698-1b homolog, and newly defined subfamilies are in red. Ten bacterial β-glucan synthase (BgsA) and fungal mixed-linkage glucan (MLG) synthase (Tft1) homologs are included to show their relationships with plant CesA/Csl subfamilies. e, Gene expression of 11 SAG 698-1b GT2 genes across 19 experimental conditions (3 replicates each). f, The phylogeny of GT2 with ZcCesA1 (Zci_04468) homologs retrieved by BLASTP against the protein non-redundant (nr) database of the National Center for Biotechnology Information (NCBI) (E-value <1 × 10−10); colors follow d, and >5,000 bacterial homologs from >8 phyla are collapsed (blue triangle). See Supplementary Data 1–13 for details. Z. cir./Z. ci, Zygnema circumcarinatum; Z. cyl., Zygnema cf. cylindricum.