Fig. 1. Open rings represent the active form of RAD52.
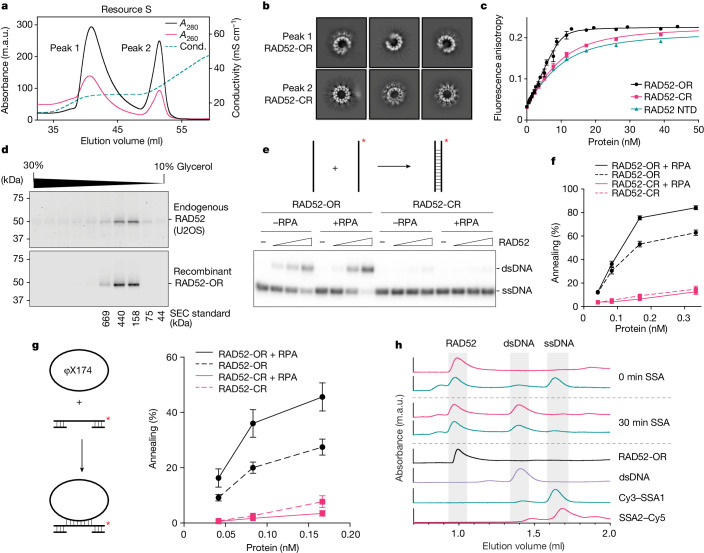
a, Resource S cation-exchange chromatography analysis of recombinant human RAD52. Cond., conductivity. b, Representative cryo-EM 2D class averages of RAD52 open (RAD52-OR) and closed rings (RAD52-CR). c, Single-stranded DNA (40 nucleotides: FAM-SSA4) binding by RAD52-OR, RAD52-CR or RAD52 NTD measured using fluorescence anisotropy. The lines are the best quadratic curve fits. Data are mean + s.e.m. n = 6 (RAD52-OR), n = 3 (RAD52-CR) and n = 3 (RAD52 NTD) independent experiments. d, Glycerol gradient sedimentation analysis of a nuclear extract from U2OS cells compared with recombinant RAD52-OR. RAD52 was detected by western blotting. Gel-filtration protein standards are shown. e, Representative PAGE assay of SSA by the open or closed rings of RAD52 (0, 0.08, 0.17 and 0.33 nM) using 68-nucleotide-long ssDNA (0.33 nM) with or without RPA (0.33 nM). f, Quantification of the SSA assays from e. Data are mean + s.e.m. n = 22 (RAD52-OR and RAD52-OR + RPA) and n = 7 (RAD52-CR and RAD52-CR + RPA) independent experiments. g, SSA using φX174 circular ssDNA and a gapped duplex by RAD52 (OR or CR) in the presence or absence of RPA. Data are mean + s.e.m. n = 4 independent experiments. h, SEC analysis of RAD52-mediated SSA between Cy3–SSA1 (dark cyan, recorded at 647 nm) and SSA2–Cy5 (pink, 555 nm) labelled ssDNAs. RAD52 was preloaded on SSA2–Cy5 before addition of Cy3–SSA1. RAD52-OR (black) was recorded at 280 nm. In e,g, 32P labels are indicated with asterisks.