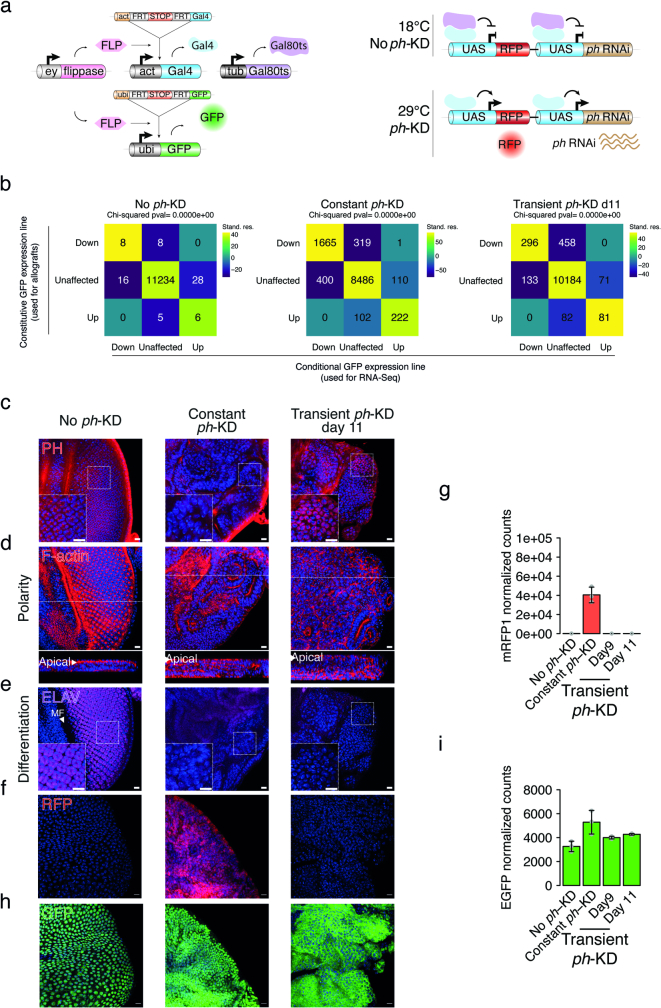
Extended Data Fig. 6. Description and validation of the conditional genetic tool allowing long-term tracking of cells subjected to constant or transient ph-KD.
a- Schematic overview of the thermosensitive ph-RNAi genetic system used in allograft experiments. Unlike the system described in Fig. 1a (conditional GFP expression), this one ubiquitously expresses GFP under the control of the Ubi-p63E promoter (constitutive GFP expression), while RFP expression is a readout of ongoing RNAi KD. b- Comparison of differentially expressed genes after no ph-KD (control), constant or transient ph-KD between the two genetic systems, allowing either the conditional (x-axis) or the constitutive (y-axis) expression of GFP. For each intersection, the corresponding number of genes is indicated (see numbers). Similarity between the two systems was assessed using chi-squared tests (see p.values on top) and chi-squared standardized residuals are shown using heat maps’ colour code. The more an intersection exceeds the size that would be expected by chance, the higher the standard residuals. c-f- PH (in red, c), F-actin (in red, d), ELAV (in magenta, e) and RFP (in red, f) stainings of EDs after no ph-KD (control, left), constant (middle) or transient ph-KD (right), respectively. PH staining was used to visualize PH loss and recovery under constant and transient conditions. F-actin is used to analyse apico-basal polarity, the neuronal marker ELAV for differentiation and RFP as a conditional marker for induction of the RNAi system. The tissues were counterstained with DAPI (blue). Two independent experiments were performed with similar results. g- Normalized read counts of mRFP1 mRNAs after no ph-KD (control), constant or transient ph-KD, showing that transcriptional expression of RFP occurs at constant 29 °C exposure but returns to basal levels after transient ph-KD. For each condition, mean normalized read counts ±standard deviation (whiskers) were inferred from three biological replicates of RNA-Seq (grey dots). h- GFP staining (in green) after no ph-KD (control), constant or transient ph-KD. GFP is constitutively expressed after transient ph-KD. The tissues were counterstained with DAPI (in blue). i- Normalized read counts of GFP mRNAs after no ph-KD (control), constant or transient ph-KD, showing that GFP expression is irreversibly induced after transient ph-KD. For each condition, mean normalized read counts ±standard deviation (whiskers) were inferred from three biological replicates of RNA-Seq (grey dots). Scale bars: 10 μm (c, d, e, f, h).