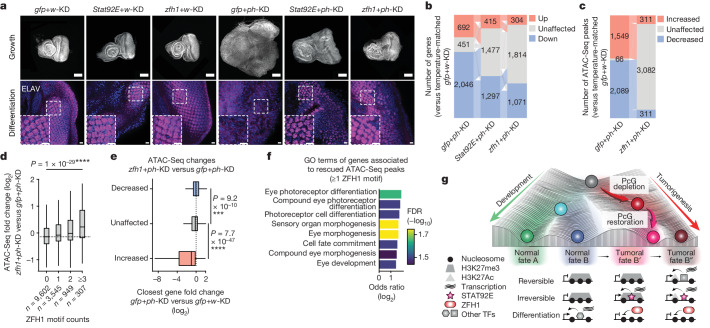
Fig. 5. Tumour development requires STAT92E and ZFH1.
a, DAPI (top, in grey) and neuronal differentiation marker ELAV (bottom, in magenta) stainings of EDs after constant KD of the following components: gfp+w, Stat92E+w, zfh1+w, gfp+ph, Stat92E+ph and zfh1+ph (top labels). Two independent biological replicates were performed with similar results. Scale bars: 100 μm (DAPI), 10 μm (ELAV). b, Number of differentially expressed genes after gfp+ph-KD (tumours), Stat92E+ph-KD and zfh1+ph-KD. Transitions between upregulated (orange), unaffected (grey) and downregulated (blue) states are indicated by thin lines of the same respective colours. c, Number of ATAC-Seq peaks showing significant accessibility changes after gfp+ph-KD or zfh1+ph-KD. Transitions between increased (orange), unaffected (grey) and decreased (blue) states are indicated by thin lines of the same respective colours. d, Fold changes at ATAC-Seq peaks between zfh1+ph-KD and gfp+ph-KD, depending on the number of ZFH1 motifs they contain (x axis). Two-sided Wilcoxon test, ****P < 1 × 10−5. Box plots show the median (line), upper and lower quartiles (box) ±1.5× interquartile range (whiskers), outliers are not shown. e, RNA-seq fold changes on gfp+ph-KD (x axis) of genes associated with ATAC-Seq peaks that are decreased (in blue), unaffected (in grey) or increased (in orange) after zfh1+ph-KD compared to gfp+ph-KD (y axis). Two-sided Wilcoxon test: ****P < 1 × 10−5. Box plots show the median (line), upper and lower quartiles (box) ±1.5× interquartile range (whiskers), outliers are not shown. f, Top enriched Gene Ontology (GO) terms for genes associated with ATAC-Seq peaks containing at least one ZFH1 motif and showing significantly increased accessibility after zfh1+ph-KD compared to gfp+ph-KD. g, Schematic illustration showing that PcG depletion triggers an epigenetic switch to a cancer fate. Resulting cancers persist after the PcG protein is restored, and their maintenance is associated with stable transcriptional changes supported by the STAT92E activator and the ZFH1 repressor.