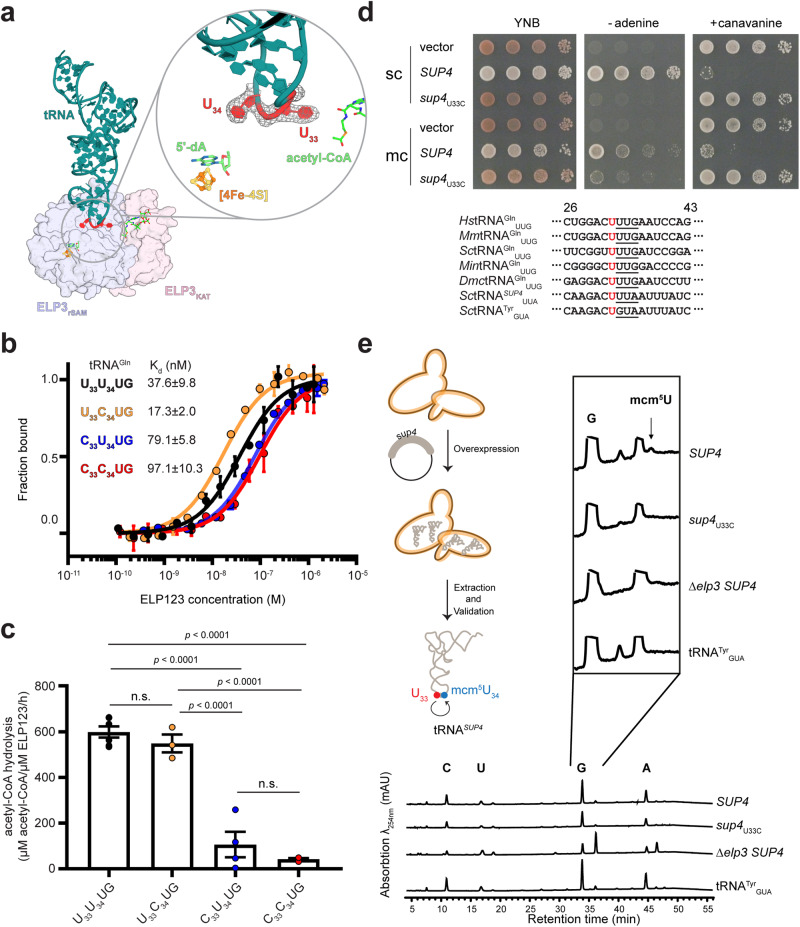
Fig. 4. Biochemical and functional characterizations of U33 and its role for U34 modification.
a Cartoon representations of tRNA bound to ELP3 and close-up view of the ASL where U33 and U34 are highlighted. The rSAM and KAT domains of ELP3 are color-coded. b MST measurements with calculated Kd values for ELP123 bound to variants of tRNAGln. n = 3 (independent experiments). Data are presented as mean values ± SEM. c Acetyl-CoA hydrolysis rates of ELP123 in the presence of variants of tRNAGln. n = 3 (independent experiments). Statistical analysis: one-way ANOVA with Tukey’s multiple comparisons test. Statistically significant differences are indicated (n.s.: not significant). Data are presented as mean values ± SEM. d Top: Phenotypic analyses of various yeast strains without or with overexpression of tRNASUP4 and tRNAsup4U33C using UAA ochre nonsense readthrough assays (sc: single copy; mc: multicopy). Elongator-modified SUP4 produces canavanine sensitivity and adenine prototrophy. Bottom: Sequence alignments of the ASL region in tRNAGln from various species as well as yeast tRNATyr and tRNASUP4. The invariant U33 is highlighted while the anticodon is underlined. e HPLC analysis of modified tRNA nucleosides. tRNA was extracted from various yeast strains expressing tRNATyrGUA, tRNASUP4, and tRNAsup4U33C. The Δelp3 deletion strain is used as a control. The peak of mcm5U is indicated by the arrow. Source data are provided as a Source Data file.