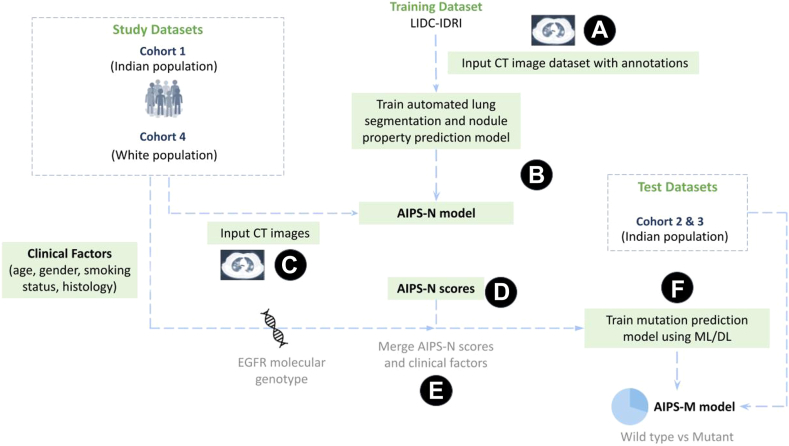
Fig. 1.
Workflow of the proposed AIPS and study design. (A) The LIDC-IDRI image and annotation dataset is downloaded from The Cancer Imaging Archive (TCIA) (B) AIPS-Nodule (AIPS-N) automated lung segmentation and nodule property prediction model is trained using the LIDC-IDRI image and annotation dataset. (C) CT images from study datasets of the Indian and White populations are fed into the trained AIPS-N model. (D) AIPS-N scores for different nodule features are calculated for each of the study datasets. (E) The AIPS-N scores, EGFR molecular genotype, and the clinical factors of the study datasets are merged. (F) Merged data is used for building the AIPS-Mutation prediction (AIPS-M) ML and DL models to ultimately predict the mutational status (wild-type or mutant) of the EGFR gene.