Figure 2.
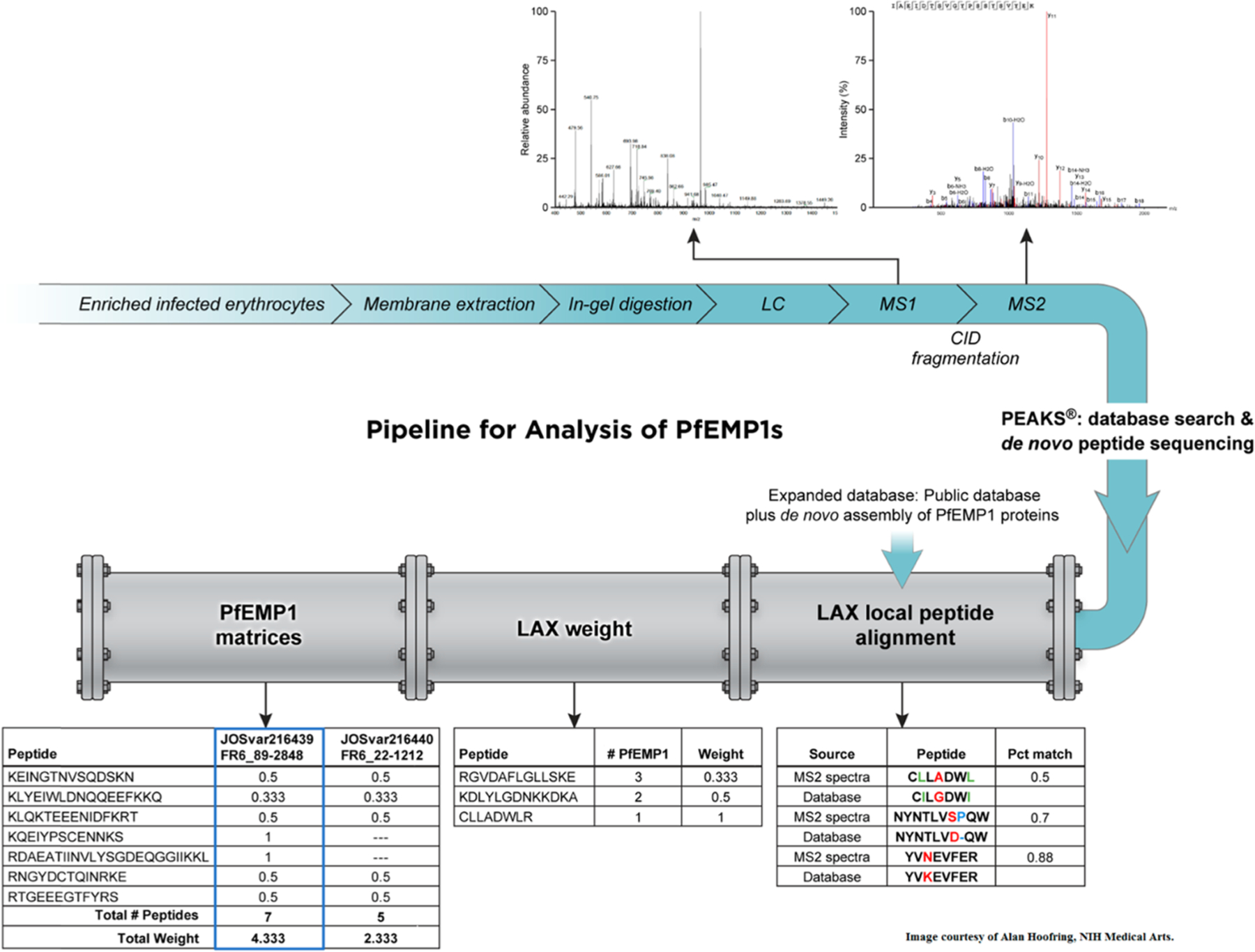
Pipeline for the analysis of PfEMP1s consists of two components: (1) Expanded Database, which contains a public database and a de novo assembly of PfEMP1 proteins. (2) The Local Peptide Alignment (LAX) algorithm, which calculates peptide weight (LAX weight) and output PfEMP1 matrices to remove redundancies. The upstream part of the figure, in blue, shows the sample preparation steps prior to analysis with the pipeline. PEAKS database search and de novo peptide sequencing results are the input for the pipeline (Image courtesy of Alan Hoofring, NIH Medical Arts).
