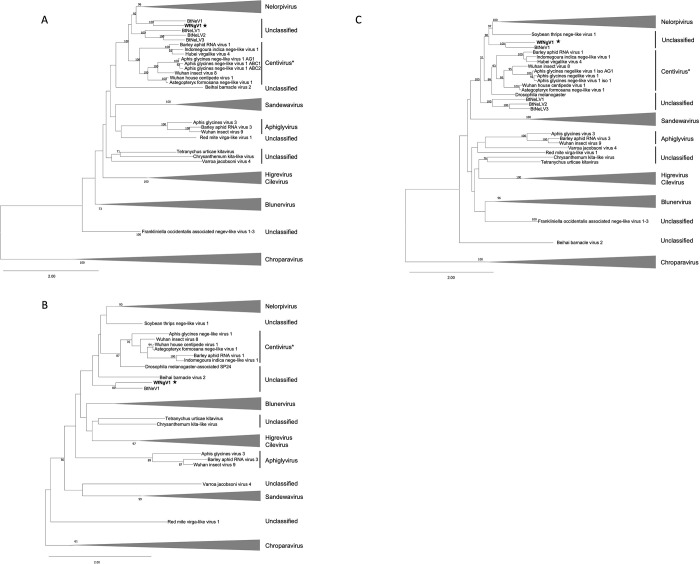
Fig 3. Maximum likelihood phylogenetic trees depicting the relationships between whitefly negevirus 1 (WfNgV1) and representative nege/kita viruses sourced from GenBank.
The analysis utilized the amino acid sequences of the RNA-dependent-RNA-polymerase A), the structural protein of 24kDa, SP24 B), and the concatenation of both (C). The LG+G+I model served as the amino acid substitution model for all analyses. The tree is drawn to scale with branch lengths measured in the number of amino acid substitutions per site. Node values represent bootstrap percentages (only values above 50% are displayed). Each taxon is labeled with the virus name, the proposed group, or as unclassified. The star indicates the WfNgV1 placement in the tree. *Additional viruses, otherwise considered unclassified such as Indomegoura indica nege-like virus 1 and Astegopteryx formosana nege-like virus 1, are included in the Centivirus due to strong bootstrap support. The complete list of virus sequences along with their accession numbers is provided in S3 Table.