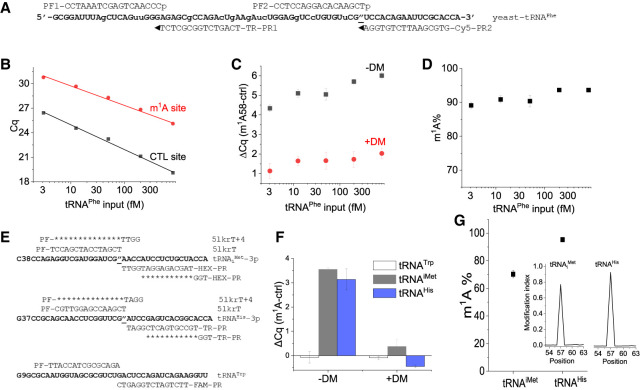
FIGURE 4.
Measuring m1A58 modification in biological RNA. (A) The sequence of yeast tRNAPhe and oligos used in TL-qPCR for m1A58 modification (″: bold and underlined). The two pairs of linkers for yeast tRNAPhe-58m1A and for -5p (control) contain sequences for Cy5 and TR (Texas Red) probe binding sites for qPCR. (B) Cq values of m1A (red) or control (black) ligation oligos of serial diluted inputs of yeast tRNAPhe from 3.1 to 800 fM without AlkB treatment. (C) Differential Cq values between m1A and control ligation oligos of serial diluted yeast tRNAPhe samples without (black) and with (red) AlkB demethylase (DM) treatment. (D) Fraction of m1A58 in yeast tRNAPhe as measured by TL-qPCR using varying amounts of input RNA. (E) Oligonucleotides and linkers used in SplintR ligation for the m1A58 site of human tRNAiMet and tRNAHis, tRNATrp is used as an internal control. Three-color qPCR is run using either HEX(m1A-iMet) + TR(m1A-His) + FAM(Trp) or HEX(CTRL-iMet) + TR(CTRL-His) + FAM(Trp) combination. (F) ΔCq values of 5lkrT (m1A) and 5lkrT + 4 (CTRL) on tRNAiMet (HEX probe), tRNAHis (TR probe), and tRNATrp (FAM probe) without (DM−) and with AlkB treatment (DM+). (G) m1A58 fraction measured by TL-qPCR for tRNAiMet and for tRNAHis. Average of n = 3 biological replicates. Inset: modification index (MI) around the m1A58 (±5 nt) site measured by DM-tRNA-seq for tRNAiMet and tRNAHis (sequencing data from NCBI GEO GSE66550).