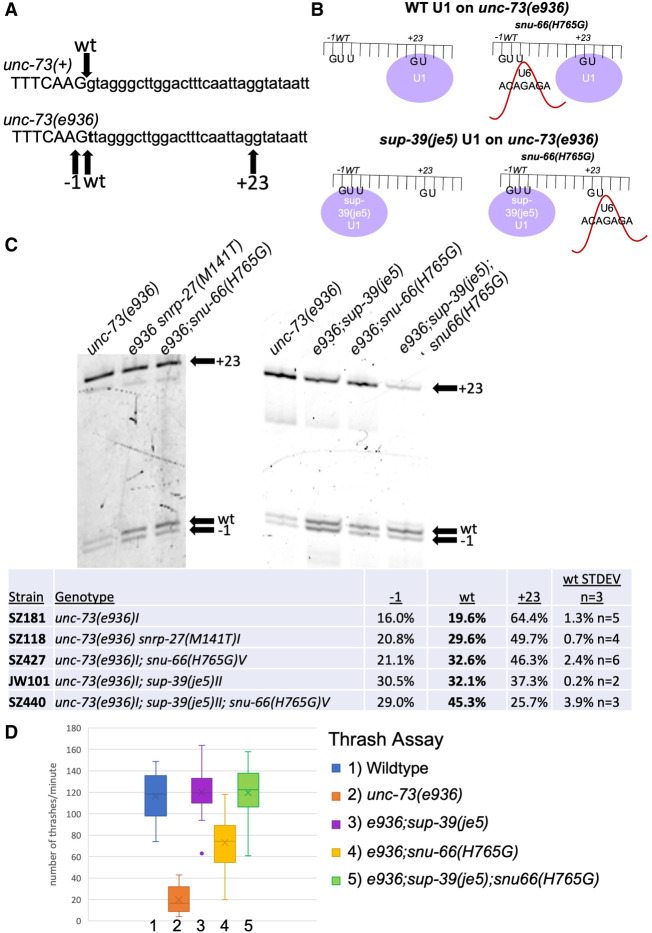
FIGURE 6.
(A) Sequences of the wt (top) and e936 mutant allele (bottom) at the start of intron 16 of the unc-73 gene. The g → t mutation is indicated for e936 in bold. The arrow above points to the wt 5′ss used in unc-73(+), and the arrows below point to the −1, wt, and +23 cryptic 5′ss activated in the unc-73(e936) mutant. (B) Cartoon representation of the hypothesis we are testing of premature U6 interaction with a weak 5′ss while U1 snRNA is still bound to a nearby 5′ss. The top part shows binding of U1 snRNA preferentially to the unc-73(e936) + 23 cryptic 5′ss on the left, and on the right side, we propose premature binding of U6 ACAGAGA region to the upstream cryptic splice sites in the presence of the snu-66(H765G) mutant. The bottom shows the U1 mutant sup-39(je5) which promotes splicing at introns that begin with UU occupying the upstream cryptic 5′ss. On the bottom right is a hypothetical outcome of splicing under these conditions, of U6 ACAGAGA interacting with the downstream 5′ss when mutant U1 occupies the upstream 5′ss. (C) Cryptic splicing of the unc-73 gene was measured using cy3 RT-PCR and separation on 6% denaturing polyacrylamide gels. The strains tested are indicated at the top and the +23, wt, and −1 splice sites are indicated on the right side of each part. Quantitation of the relative usage of the three cryptic splice sites is shown below the gel images. (D) Box and whisker chart representing the thrash test data to demonstrate phenotypic suppression of unc-73(e936) uncoordination. The y-axis represents the average number of times a worm bent across their body axis in 1 min; 20 L4 worms were assayed for each strain.