Figure 4.
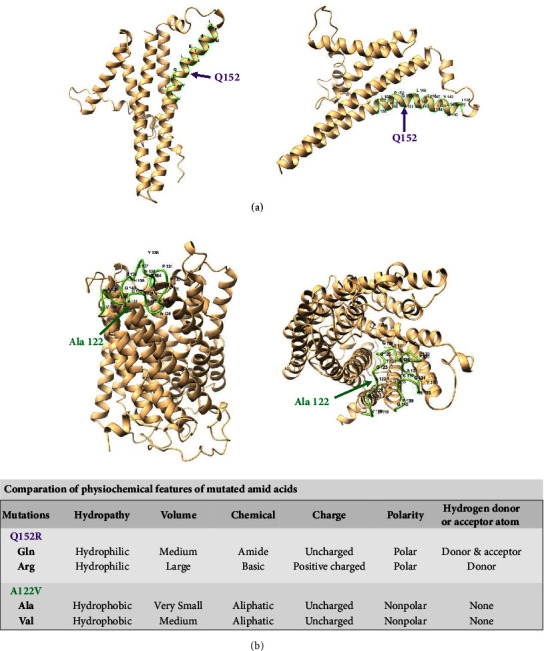
Structural localization of identified ChrMT DNA mutations in proteins and analyses of physiochemical features of mutated amino acids. (a) Complex V subunit A and ChrMT: 8981A > G (ATP6: Q152R, pointed by purple arrows). The mutation is located in one transmembrane helical structure (138–158, highlighted in green). The mutation from Gln to Arg changes certain physiochemical features. (b) Complex IV subunit 1 and ChrMT: 6268C > T (Cox1: A122V, pointed by green arrows). The mutation from Ala to Val is located in a topological domain (118–140, highlighted in green) and appears to cause less consequence on the protein structure and/or function. Structure predictions were prepared using AlphaFold [34].
