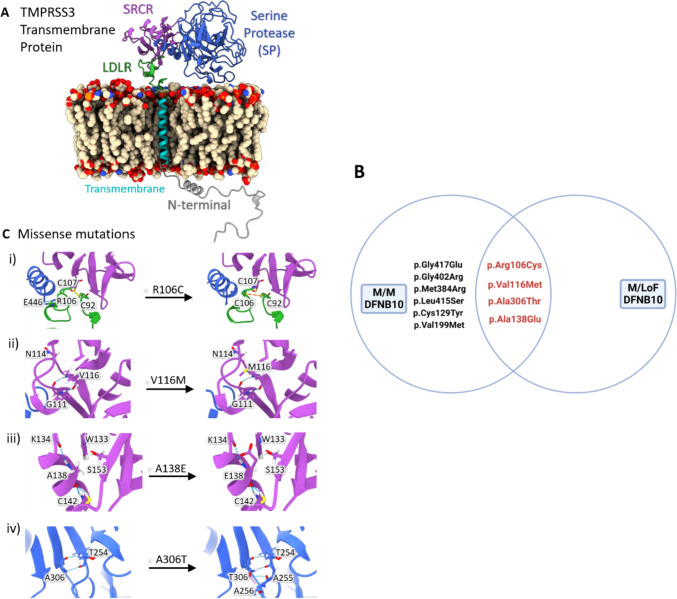
Fig. 5.
Protein modeling shows deleterious affects of 4 missense mutations. A Human TMPRSS3 model predicted by AlphaFold2, positioned in a lipid bilayer generated by Charmm-gui. The 4 domains of the protein are highlighted and labeled. B Overlap of missense variants that lead to severe hearing loss or M/WT with clinical hearing loss. The consistent overlap suggests these 4 variants are more severe in their effects. C Zoomed-in structures showing the differences in interactions due to these 4 missense mutations i) R106C mutation showing the potential disulfide bond formation in yellow dotted lines between C106 and C92. ii) V116M showing the clashing of the mutant Methionine with N114. iii) A138E shows the insertion of the large negatively charged Aspartate residue, inducing steric clashes with nearby amino acids W133, K134, and S153. iv) A306T showing two extra backbone hydrogen bonds formed by the mutant methionine residue with A255 and A256