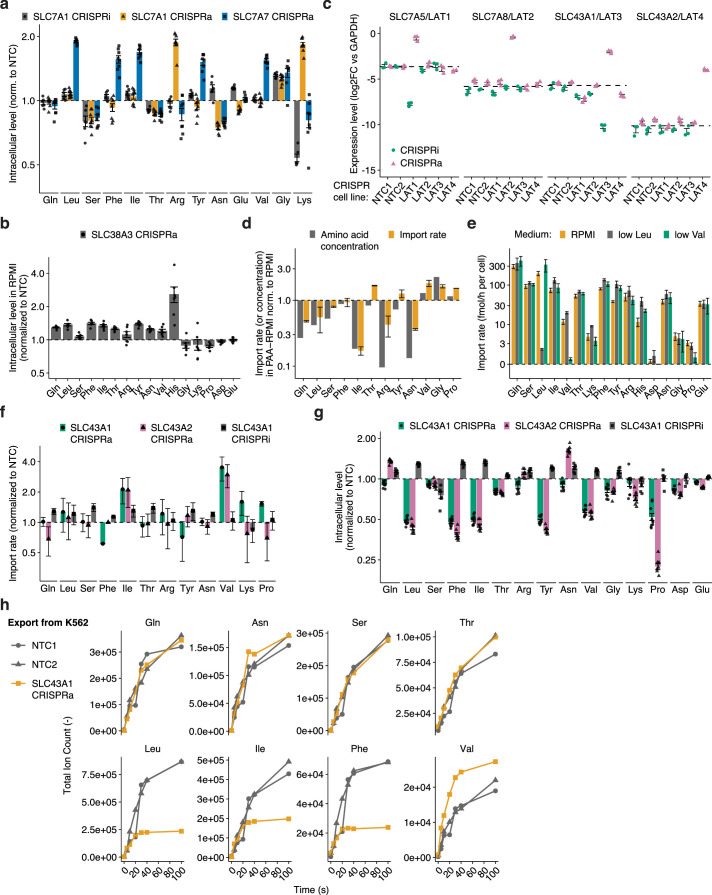
Extended Data Fig. 5. Specific changes to amino acid levels and transport rates induced by CRISPRi/a of SLCs.
(a) Changes in intracellular amino acid levels induced by CRISPRi/a of SLC7A1 and SLC7A7. (b) Same as (a) but for SLC38A3 CRISPRa and data from Fig. 3d. (c) LAT1–4 expression levels in K562 CRISPRi/a cell lines were determined by RT-qPCR relative to the housekeeping gene GAPDH. n = 3 technical replicates. Data are mean ± s.e.m. (d) Amino acid import rates into K562 cells correlate with levels of amino acid in the growth medium. Import rates for K562 CRISPRa SLC43A1 and non-targeting control (NTC) in RPMI and PAA–RPMI were from Fig. 3f. Data represent the ratio of import rates in PAA–RPMI to that in RPMI. Relative amino acid levels in the media were determined from their respective formulation. (e) The import of amino acids into K562 cells in low amino acid medium is selectively diminished for that specific low abundance amino acid. Data: mean ± s.e.m. of n = 2 (low Val), n = 3 (low Leu), n = 4 (RPMI) independent import rate determinations each calculated from the linear regression of n = 6 biologically independent samples. (f) SLC43A2 CRISPRa increases import of isoleucine and valine into K562 cells in RPMI. Rates were determined from a linear regression of n = 6 biologically independent samples. Data represent the slope ± SE normalized to NTC. Data for SLC43A1 CRISPRa is from Fig. 3f. (g) SLC43A2 CRISPRa induces a decrease in intracellular levels of large neutral amino acids in K562 cells grown in RPMI. (h) SLC43A1 CRISPRa leads to higher export of valine and similar export of leucine, isoleucine, and phenylalanine despite lower intracellular pools. Cells grown in RPMI containing heavy-isotope labelled amino acids were rapidly washed, then incubated in regular RPMI. Accumulation of heavy-labelled amino acids in RPMI was monitored over time by GC-MS. Representative example of 3 independent experiments. (a,b,d,g) n = 7 biologically independent samples. Data are mean ± s.e.m. Source numerical data are available in source data.