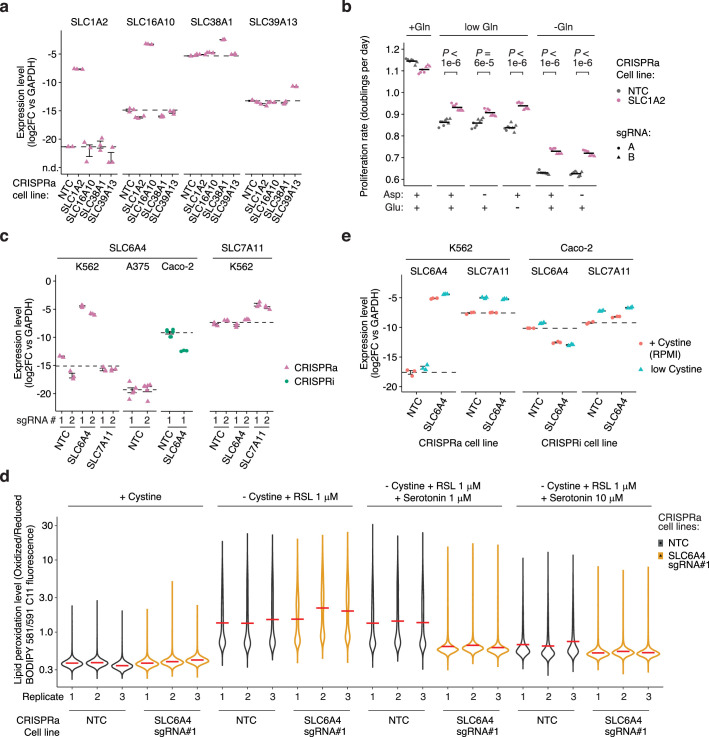
Extended Data Fig. 6. Specific changes to expression level of SLCs by CRISPRi/a.
(a) RT-qPCR analysis shows strong and specific gene upregulation by CRISPRa. n = 3 technical replicates. Data are mean ± s.e.m. (b) SLC1A2 CRISPRa confers a growth advantage in low and no glutamine RPMI in the presence or absence of either glutamate or aspartate in the medium. Proliferation rates were extracted from competition assays with 4 biologically independent samples from two independent sgRNAs. P values were determined using two-tailed unpaired Student’s t-tests. (c) Expression level of SLC6A4 in cell lines used in this study and specific up- and down-regulation of SLC6A4 via CRISPRi/a. Levels of SLC6A4 and SLC7A11 were quantified by RT-qPCR relative to the housekeeping gene GAPDH in K562 CRISPRa, A375 CRISPRa, and Caco-2 CRISPRi cell lines with sgRNAs targeting SLC6A4, SLC7A11 or a non-targeting control (NTC). n = 3 technical replicates, except for A375 and Caco-2 NTC where n = 6. Data are mean ± s.e.m. (d) Violin plots displaying the distribution of lipid peroxidation levels in single cells (~20k) as determined by flow cytometry of cells incubated with BODIPY 581/591 C11 sensor after growth in the mentioned conditions. Red bars represent the average peroxidation level of the population and were used in Fig. 4g. (e) Low cystine induces expression of SLC7A11 but not of SLC6A4. K562 and Caco-2 cells were grown in either RPMI or low cystine RPMI for 3 days with daily media changes. Expression levels were quantified by RT-qPCR relative to GAPDH (n = 3 technical replicates. Data are mean ± s.e.m.). Source numerical data are available in source data.