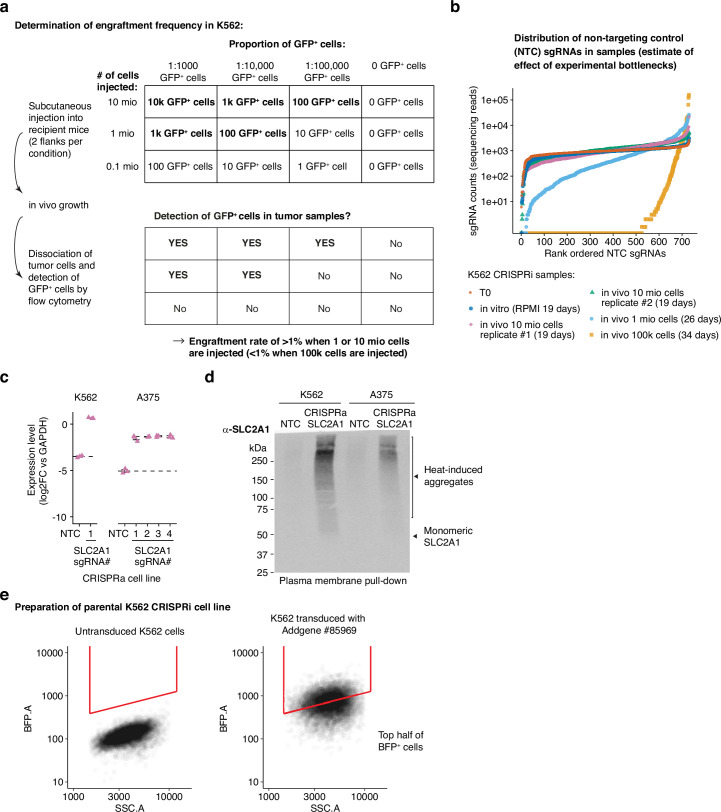
Extended Data Fig. 8. Optimization of subcutaneous CRISPRi/a transporter screens.
(a) Determination of the engraftment frequency of K562 cells injected subcutaneously into immunodeficient mice. K562 CRISPRi library cells expressing GFP were mixed into pools of K562 CRISPRi library cells at a ratio ranging from 1:1000–1:100,000. 10, 1 or 0.1 million cells of these preparations were injected into the flanks of mice. Tumors formed over 19–34 days, were harvested and dissociated into single cell suspensions. The presence (or absence) of GFP+ cells in homogenized tumor samples was determined by flow cytometry and was used to determine the engraftment frequency and conditions required to preserve library complexity during in vivo transporter screens. (b) Plot displaying the number of counts for all non-targeting control (NTC) sgRNAs determined either from the sequencing of tumor samples from (a), from the initial pool of library cells, or from library cells grown in RPMI. (c) Expression level of SLC2A1 in K562 and A375 CRISPRa cell lines determined by RT-qPCR and relative to the housekeeping gene GAPDH. n = 3 technical replicates. Data are mean ± s.e.m. (d) Overexpression of SLC2A1 via CRISPRa leads to increased levels at the plasma membrane in K562 and A375 cells. Cells were incubated with a cell-impermeable biotinylation reagent, and plasma membrane proteins were isolated by streptavidin affinity purification and analyzed by Western blotting. (e) Example of gating strategy used for flow cytometry sorting of K562 cells transduced with lentivirus expressing dCas9-BFP-KRAB for K562 CRISPRi/a cell line generation. Source numerical data and unprocessed blots are available in source data.