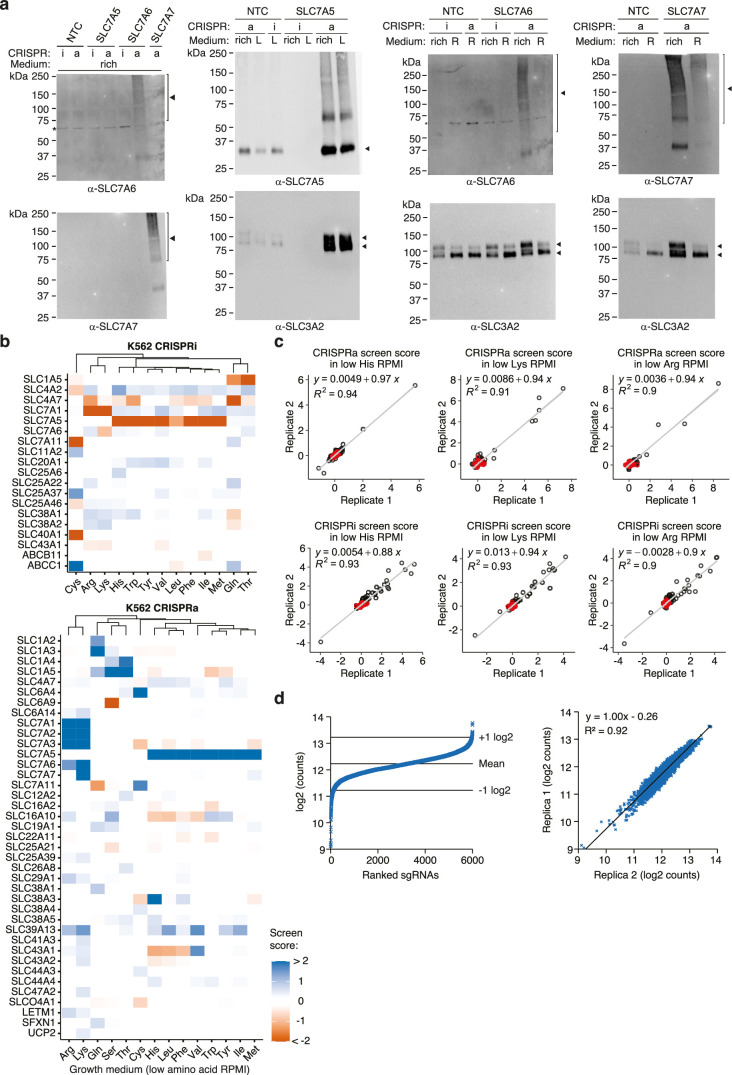
Extended Data Fig. 2. CRISPRi/a transporter screens in low amino acid conditions.
(a) CRISPRi/a of transporters leads to changes in protein level at the plasma membrane. Changes in low Leu (L) and low Arg (R) conditions are similar to those observed in RPMI (rich). K562 CRISPRi/a cells were incubated with a cell-impermeable biotinylation reagent, and plasma membrane proteins were isolated by streptavidin affinity purification and analyzed by Western blotting. * denotes a non-specific band. (b) Tile plots displaying all significant transporter CRISPRi/a hits in low amino acid screens. Screen scores were calculated by multiplying phenotype scores by -log10(P value) for all genes and computed negative controls. P values were determined using a Mann-Whitney test. A stringent score cutoff was determined by the highest scoring negative control across all conditions for both datasets. All transporter genes that were significant in at least one condition tested were included in the tile plot. Transporter CRISPRi/a cell lines that have a growth defect in RPMI were prone to being pan-resistant in low amino acid conditions, as previously reported in other screens25, and were removed from the analysis (see Methods). Columns were ordered based on hierarchical clustering of scores. (c) CRISPRi/a transporter screens are highly reproducible as shown by the strong correlation between independent screen replicates. Black circles represent individual transporter genes and red circles indicate negative control genes. (d) The custom sgRNA library is highly homogenous, and the library preparation introduces no sample bias. gDNA was isolated from a pool of K562 CRISPRi transporter library cells post-antibiotic selection, and the abundance of each sgRNA present in the library was determined after PCR amplification and high-throughput sequencing (Methods). For this representative sample, >97% of sgRNAs were within 1 log2 of the mean, 12 sgRNAs had less than 1000 counts, and no sgRNA had 0 counts. Source numerical data and unprocessed blots are available in source data.