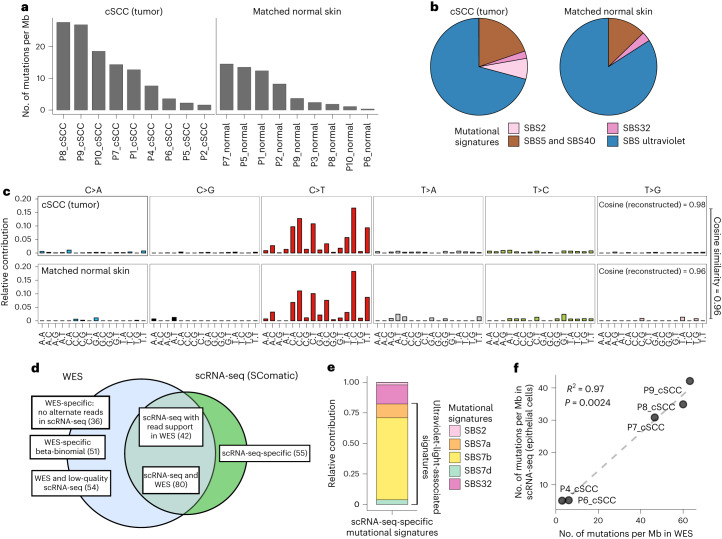
Fig. 2. Validation of SComatic using matched scRNA-seq and exome sequencing data.
a, Mutational burdens for epithelial cells using the somatic SNVs detected by SComatic in cSCC and matched normal skin scRNA-seq data sets. The number of mutations is normalized to account for the variable number of callable sites in each sample. b, Fraction of somatic SNVs detected in epithelial cells attributed to COSMIC signatures. SBS signatures associated with ultraviolet radiation (SBS7a, SBS7b, SBS7c and SBS7d) and clock-like mutational processes (SBS5 and SBS40) are collapsed for visualization purposes. c, Mutational spectra computed for the mutations detected using SComatic in epithelial cells from cSCC and matched normal skin scRNA-seq data. The cosine similarities between the observed and reconstructed mutational spectra are shown. d, Venn diagram showing the overlap of the somatic SNVs detected by SComatic in epithelial cells using scRNA-seq data and WES data from the cSCC samples. ‘WES-specific beta-binomial’ refers to mutations detected in WES with at least one alternative read count in scRNA-seq that are not significant for the beta-binomial test. e, Decomposition of the mutations detected in scRNA-seq data only (scRNA-seq-specific mutations) into COSMIC signatures. f, Correlation between the mutational burdens estimated using the mutations detected in WES and the mutations detected by SComatic in the scRNA-seq data. The correlation was assessed using a linear regression model. Only genomic regions with sufficient sequencing depth in both the WES and scRNA-seq data were considered for this analysis. Mb, megabase.