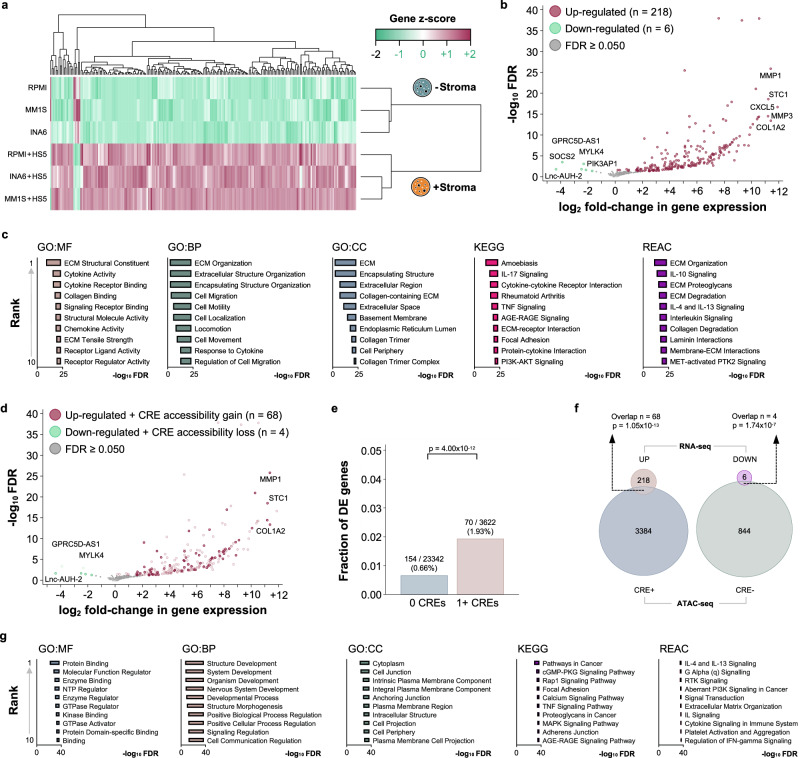
Fig. 2. The interaction of bone marrow stroma and myeloma cells induces up-regulation of genes involved in cell migration and cytokine signaling in human myeloma cell lines.
This up-regulation is supported by the presence of de novo chromatin accessibility in associated cis-regulatory elements. a Heatmap (unsupervised hierarchical clustering) showing separation of the experimental conditions based on gene expression (presence versus absence of stromal co-culture). b Volcano plot demonstrating the up-regulation of genes in myeloma cells due to interactions with bone marrow stroma. Genes coding for interleukins, chemokine ligands, and matrix metalloproteinases were among the top up-regulated genes (top 5 genes in each direction labeled). c Bar graphs listing the top 10 functional annotation terms for the 224 differentially expressed genes (GO = Gene Ontology, MF = Molecular Function, BP = Biological Process, CC = Cellular Compartment, KEGG = Kyoto Encyclopedia of Genes and Genomes Pathway, REAC = Reactome Pathway). These top 10 terms included cytokine and interleukin signaling as well as cell migration and remodeling of the extracellular matrix. d Volcano plot demonstrating differential expression of genes with concomitant differential chromatin accessibility in associated CREs. Genes coding for interleukins, chemokine ligands, and matrix metalloproteinases remained among the top up-regulated genes. The top 3 differentially expressed genes with associated differentially accessible CREs are labeled. e Bar graph showing the over-representation (Fisher’s exact test) of up-regulated genes among the genes with at least one CRE with altered accessibility nearby (i.e. genes close to CREs with altered chromatin accessibility after stromal co-culture are more likely to be up-regulated after stromal co-culture compared to genes without such CREs nearby). f Venn diagrams demonstrating that the observed association between differential expression and differential chromatin accessibility in corresponding CREs is unlikely to have arisen by chance (hypergeometric test). g Bar graphs listing the top 10 functional annotation terms for the 4626 predicted target genes predicted to be involved in cis-interactions. These top 10 terms again included interleukin and cytokine signaling as well as several cancer-related pathways. Source data are provided as a Source Data file.