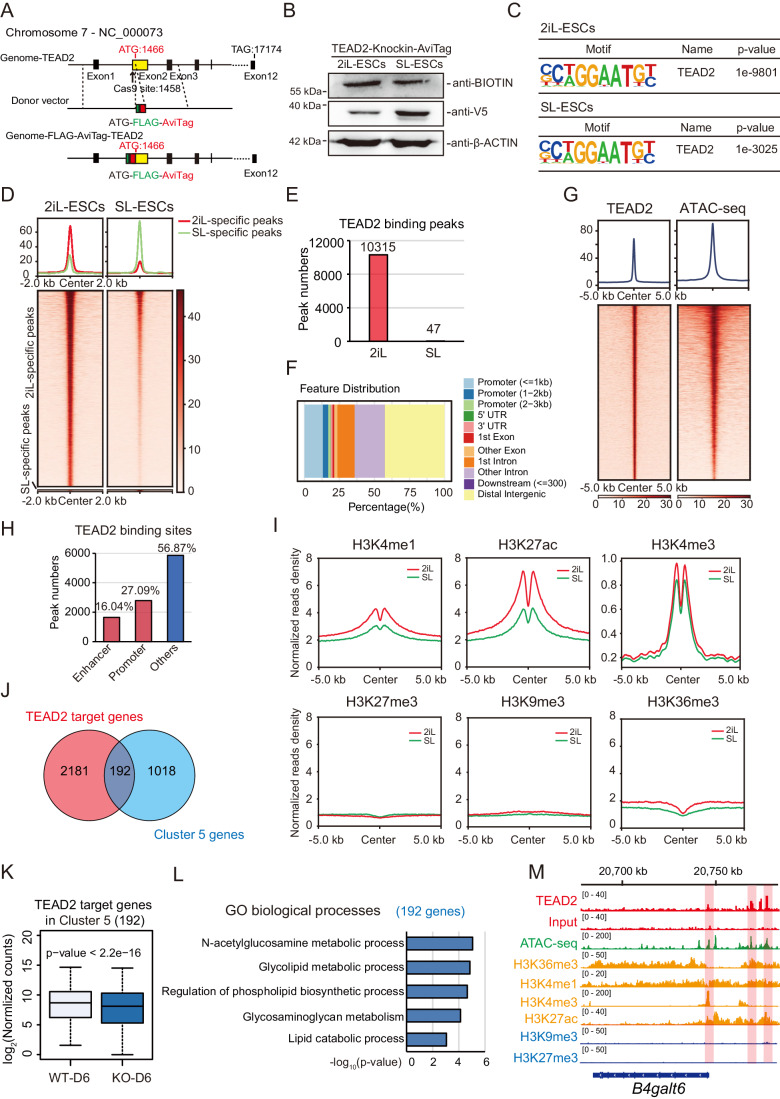
Figure 4. TEAD2 binds to the active chromatin regions of 2i-specific genes.
(A) Strategy for generating of Tead2-FLAG-AviTag knock-in cell lines in 2iL- and SL-ESCs. (B) Western blot analysis for BIOTIN and V5 with cell lysates from Tead2-FLAG-AviTag-knock-in 2iL- and SL-ESC lines. β-ACTIN was used as a loading control. (C) Motif-enrichment analysis of BIOTIN-binding sites in 2iL- and SL-ESCs. P-value was calculated by using hypergeometric enrichment calculations from Homer software. (D) Heatmap showing the comparison of TEAD2 binding sites between 2iL- and SL-ESCs. (E) Number of 2iL- and SL-ESCs-specific TEAD2 binding peaks. (F) Pie charts showing the genomic distribution of 2i-specific TEAD2 peaks. (G) Heatmaps of sequence read density for ATAC-seq in 2i-specific TEAD2 binding peaks. (H) Bar plot showing the number of 2i-specific TEAD2 peaks that overlap with both promoters and enhancers. (I) Tag-density pileup showing H3K4me1, H3K27ac, H3K4me3, H3K27me3, H3K9me3, and H3K36me3 ChIP-seq signals at the 2i-specific TEAD2 binding sites in both 2iL- and SL-ESCs. (J) Venn plots showing the overlap among 2i-specific TEAD2 target genes and cluster 5 genes. (K) Boxplots showing expression level of overlapping genes in (J) between wild-type and Tead2-knockout cells at day 6 of the transition. The centerline indicates the median value, while the box and whiskers represent the interquartile range (IQR) and 1.5 × IQR, respectively, n = 192. P-value was calculated by Mann–Whitney U test. (L) GO categories of the overlapping genes shown in (J). The enrichment p-value was calculated by using Metascape software. (M) Genomic views of enrichment for TEAD2 and histone modifications in the B4galt6 gene. Source data are available online for this figure.