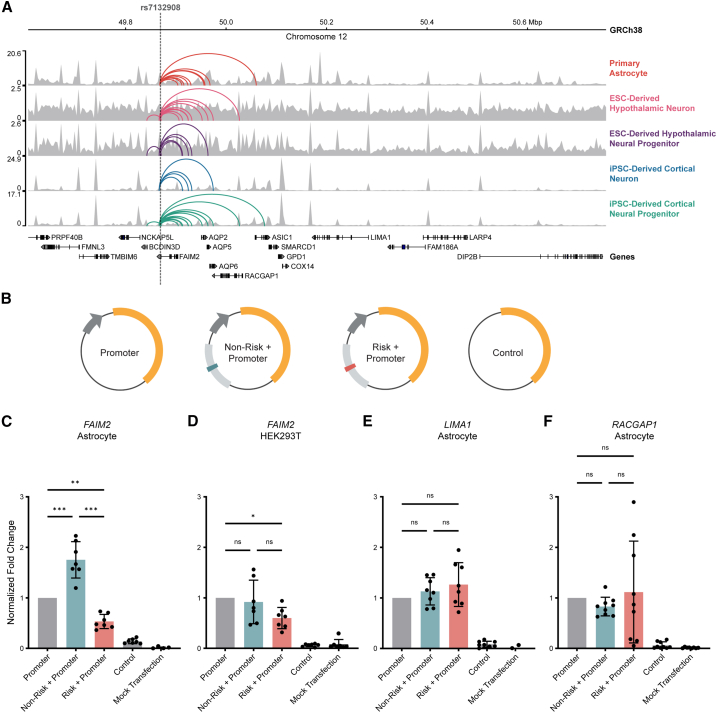
Figure 1.
rs7132908 regulates FAIM2 expression with allele and cell-type specificity
(A) Chromatin accessibility represented by ATAC-seq tracks depicting normalized reads and chromatin loops at the TAD containing rs7132908 in neural cell types. Chromatin loops represent significant contacts between regions of open chromatin that harbored rs7132908 and a gene promoter. Gray dashed vertical line indicates rs7132908 position.
(B) Graphic representation of firefly luciferase reporter vectors used in luciferase reporter assays.
(C‒F) Fold change of firefly luciferase fluorescence normalized to the promoter only control vector driven by the FAIM2 promoter in primary astrocytes (n = 7 biological replicates) (C), FAIM2 promoter in HEK293Ts (n = 7 biological replicates) (D), LIMA1 promoter in primary astrocytes (n = 8 biological replicates) (E), and RACGAP1 promoter in primary astrocytes (n = 9 biological replicates) (F). Data are represented as mean ± SD. ∗p value <0.05, ∗∗p value <0.01, ∗∗∗p value <0.001 by one-way ANOVA with Tukey’s correction for multiple comparisons.