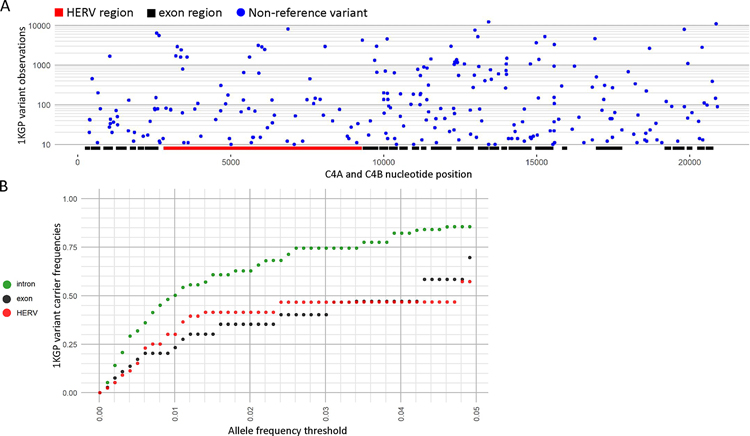
Figure 3. SNV variation across the 1KGP dataset.
(A) Total copy of combined C4A and C4B non-reference variants, which are variants not represented in the main assembly of GRCh38, by C4A and C4B position for the 1KGP dataset. The copy number of all non-reference variants for a position across the 1KGP dataset are summed to get the non-reference variant copy, which was then filtered to only show variant positions with total copy of at least 10. Positions of exon and HERV-K(C4) regions are marked. (B) Global carrier frequencies for non-reference variants in the 1KGP dataset for increasing global allele frequency thresholds from 0.00–0.05 for introns, exons, and the HERV-K(C4) region. The y-axis represents the total proportion of carriers that carry a non-reference allele that is at or below the global allele frequency threshold on the x-axis. For example, nearly 25% of the 1KGP dataset carried exonic variants with a global allele frequency of 1% or lower.