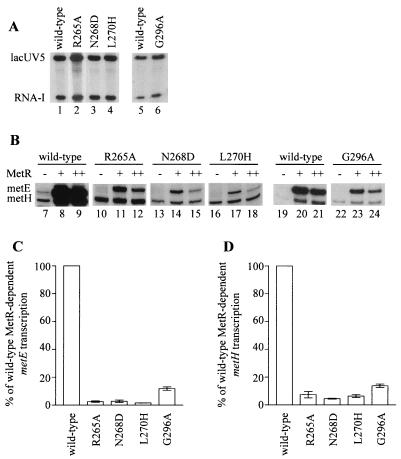
FIG. 3.
In vitro transcriptions using RNAP with αCTD substitutions. The RNAP is designated by the α subunits in the holoenzyme. (A) Transcripts from single-round transcription experiments with the supercoiled plasmid pRLG593 as a template to equalize the activity of each RNAP. Indicated are the positions of the αCTD-independent transcript initiated at the lacUV5 promoter and the plasmid-derived RNA-I transcript. The activity of the G296A RNAP was determined in a separate experiment, so it is shown with the wild-type RNAP from the same experiment. RNAPs were used at the following concentrations in these experiments: 20 nM wild-type, R265A, and N268D RNAPs, 60 nM L270H RNAP, and 50 nM G296A RNAP. (B) Transcripts from single-round transcription experiments using a linear template carrying the metE-metR control region as well as a linear template with the metH promoter in the absence (−) and the presence of 135 nM (+) or 270 nM (++) MetR dimer. The transcripts initiated at the metE and metH promoters are indicated. (C and D) Effects of αCTD substitutions on metE (C) and metH (D) activation. The metE transcript produced by each RNAP in the presence of 135 nM MetR and the metH transcript produced by each RNAP in the presence of 270 nM MetR from two independent in vitro transcription experiments were quantitated and normalized to the amounts of lacUV5 and RNA-I transcripts produced by the same RNAP. The normalized amount of transcript produced by each mutant RNAP is reported as a percentage of the expression by wild-type RNAP (± 1 standard deviation).