Figure 3. Functional profiles at the D3R.
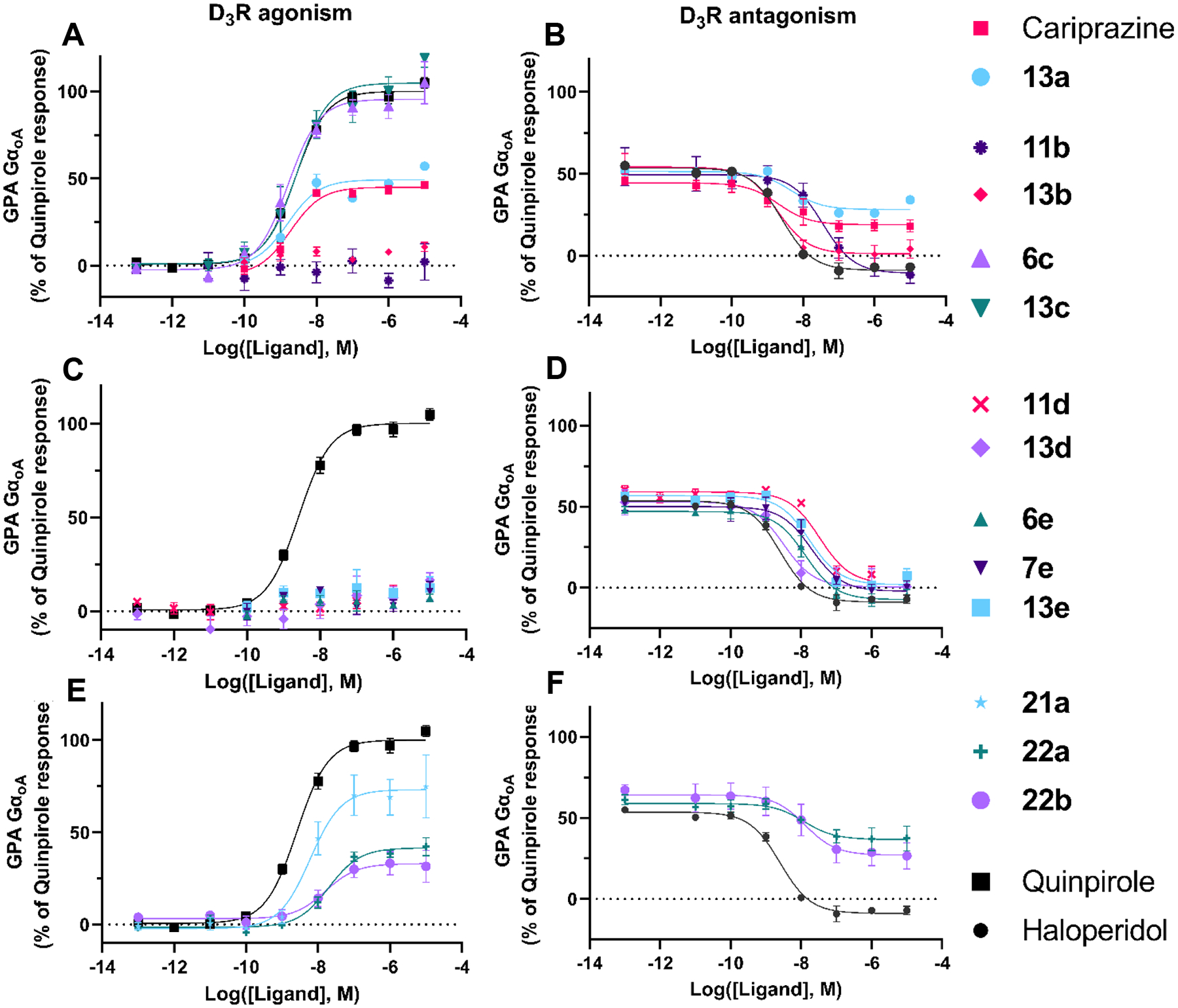
Using the GαoA assay, all ligands were tested for D3R agonism, shown on the left panels and 11 out of the 14 selected ligands were tested for D3R antagonism (compounds were added to cells together with and EC50 concentration (3 nM) of quinpirole), shown on the right panels. 6c, 13c, and 21a signaled as robust D3R agonists and were thus not tested as antagonists as they would not displace quinpirole signal. Each panel shows concentration response curves for a subset of the selected ligands: 2,3-dichlorophenyl, 2-chloro-3-ethylphenyl, and 2-fluoro-3-methoxyphenyl compounds (A) D3R agonism and (B) D3R antagonism; 2-trifluoromethyl substituted pyridine and 3-chloro-5-ethyl-2-methoxyphenyl compounds (C) D3R agonism and (D) D3R antagonism; linker modified compounds (E) D3R agonism and (F) D3R antagonism. Data points represent the mean ± SEM of three independent experiments performed in duplicate.
